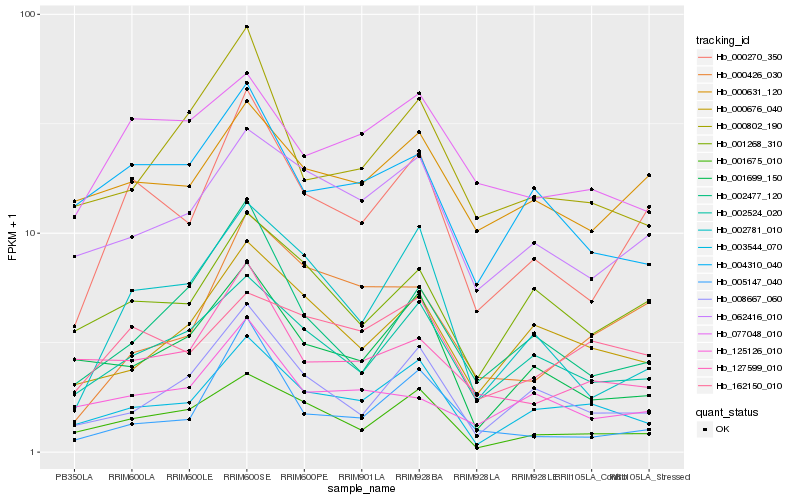
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_350 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 2 |
Hb_002524_020 |
0.1470388183 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_002781_010 |
0.1485331683 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_062416_010 |
0.1545532795 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 5 |
Hb_001268_310 |
0.1561282839 |
- |
- |
- |
| 6 |
Hb_001675_010 |
0.1569473764 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 7 |
Hb_003544_070 |
0.1632455409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000631_120 |
0.1653793927 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 9 |
Hb_002477_120 |
0.1656923182 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_008667_060 |
0.1701213859 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 11 |
Hb_000426_030 |
0.1736492487 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004310_040 |
0.1739295829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001699_150 |
0.1775703784 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 14 |
Hb_125126_010 |
0.1780717696 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 15 |
Hb_127599_010 |
0.1781371199 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 16 |
Hb_162150_010 |
0.1784692748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005147_040 |
0.1789102081 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 18 |
Hb_000802_190 |
0.1797730458 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 19 |
Hb_000676_040 |
0.1803973841 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 20 |
Hb_077048_010 |
0.1819373633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |