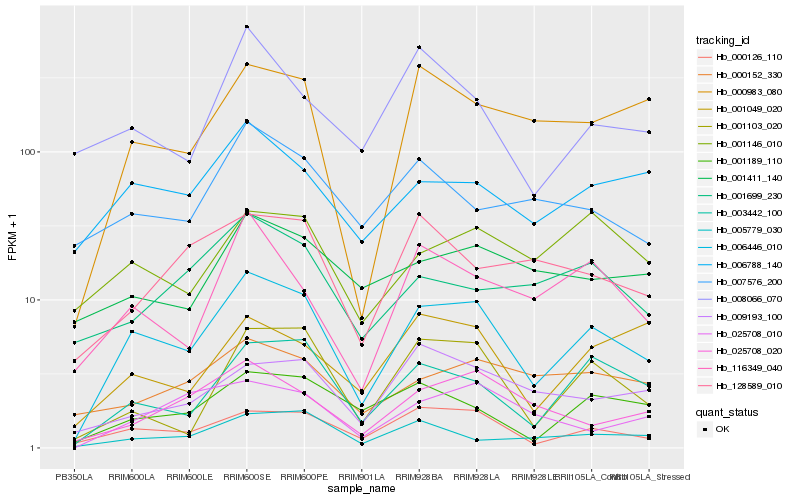
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006446_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000126_110 |
0.1383460767 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 3 |
Hb_003442_100 |
0.1549268965 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 4 |
Hb_001103_020 |
0.1808567977 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 5 |
Hb_000983_080 |
0.1817393419 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 6 |
Hb_006788_140 |
0.1882230811 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 7 |
Hb_001189_110 |
0.1882967711 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 8 |
Hb_116349_040 |
0.1926804163 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 9 |
Hb_001146_010 |
0.1998461666 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 10 |
Hb_025708_020 |
0.2004272616 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005779_030 |
0.2021888644 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 12 |
Hb_000152_330 |
0.2042532306 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 13 |
Hb_025708_010 |
0.2045352996 |
- |
- |
hypothetical protein MTR_1g051060 [Medicago truncatula] |
| 14 |
Hb_001049_020 |
0.2065062685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009193_100 |
0.2070282094 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 16 |
Hb_001699_230 |
0.2087439641 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 17 |
Hb_008066_070 |
0.2090300927 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 18 |
Hb_128589_010 |
0.2100244871 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 19 |
Hb_001411_140 |
0.2110220336 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 20 |
Hb_007576_200 |
0.2116186226 |
- |
- |
amino acid transporter, putative [Ricinus communis] |