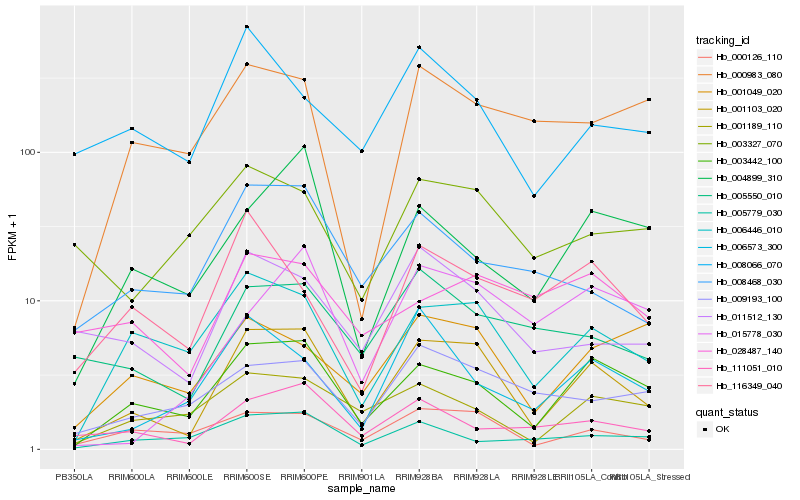
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001103_020 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 2 |
Hb_003442_100 |
0.1601867403 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 3 |
Hb_006446_010 |
0.1808567977 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000126_110 |
0.1839859955 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 5 |
Hb_111051_010 |
0.220975893 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 6 |
Hb_011512_130 |
0.2226306757 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 7 |
Hb_001189_110 |
0.2263351765 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 8 |
Hb_005550_010 |
0.2272117973 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 9 |
Hb_009193_100 |
0.2286723794 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 10 |
Hb_006573_300 |
0.2320201684 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_008468_030 |
0.2360533689 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 12 |
Hb_001049_020 |
0.2378238437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004899_310 |
0.2401164273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_116349_040 |
0.2419244226 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 15 |
Hb_003327_070 |
0.2432138726 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 16 |
Hb_008066_070 |
0.2433268016 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 17 |
Hb_015778_030 |
0.2457782207 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 18 |
Hb_000983_080 |
0.245922929 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 19 |
Hb_005779_030 |
0.247188707 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 20 |
Hb_028487_140 |
0.252819973 |
- |
- |
PREDICTED: myosin-binding protein 2 [Jatropha curcas] |