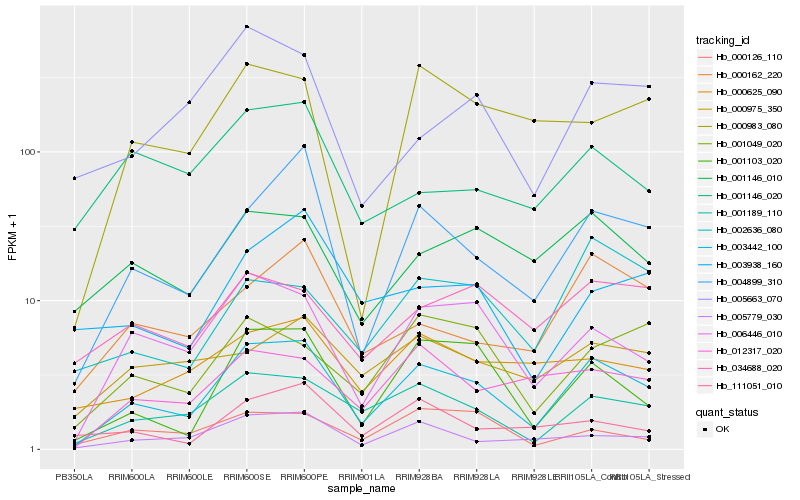
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003442_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 2 |
Hb_001189_110 |
0.13796607 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 3 |
Hb_006446_010 |
0.1549268965 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_004899_310 |
0.1559620761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001103_020 |
0.1601867403 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 6 |
Hb_005779_030 |
0.1736603433 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 7 |
Hb_012317_020 |
0.1906213229 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_001146_010 |
0.1924327755 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 9 |
Hb_000162_220 |
0.1941744431 |
- |
- |
nodulin family protein [Populus trichocarpa] |
| 10 |
Hb_001049_020 |
0.1976178726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000625_090 |
0.2016813757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000983_080 |
0.202127958 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 13 |
Hb_000975_350 |
0.2047748715 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 14 |
Hb_111051_010 |
0.2050232882 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 15 |
Hb_005663_070 |
0.2056502726 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 16 |
Hb_034688_020 |
0.2060828394 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 17 |
Hb_000126_110 |
0.206670935 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 18 |
Hb_003938_160 |
0.209000182 |
- |
- |
PREDICTED: transcription factor bHLH155 [Jatropha curcas] |
| 19 |
Hb_002636_080 |
0.2123590648 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 20 |
Hb_001146_020 |
0.2129730643 |
- |
- |
hypothetical protein CISIN_1g013079mg [Citrus sinensis] |