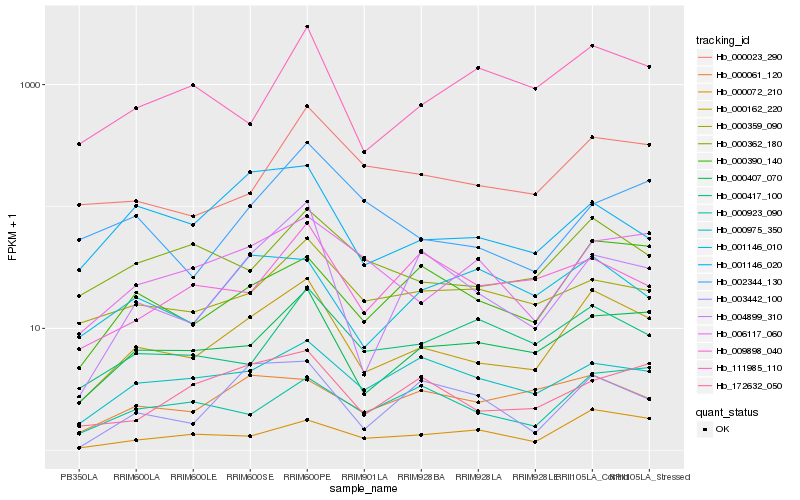
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_220 |
0.0 |
- |
- |
nodulin family protein [Populus trichocarpa] |
| 2 |
Hb_000407_070 |
0.1369711456 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000390_140 |
0.1665486515 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 4 |
Hb_000023_290 |
0.174629789 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000362_180 |
0.1749623648 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 6 |
Hb_006117_060 |
0.1769541126 |
- |
- |
hypothetical protein CARUB_v10024452mg [Capsella rubella] |
| 7 |
Hb_000072_210 |
0.177289878 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 8 |
Hb_004899_310 |
0.1799110542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000975_350 |
0.1833885399 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 10 |
Hb_172632_050 |
0.1837039486 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 11 |
Hb_001146_020 |
0.1851103692 |
- |
- |
hypothetical protein CISIN_1g013079mg [Citrus sinensis] |
| 12 |
Hb_000359_090 |
0.1878434715 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 13 |
Hb_003442_100 |
0.1941744431 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 14 |
Hb_000417_100 |
0.1953385879 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 15 |
Hb_111985_110 |
0.1953671742 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 16 |
Hb_000061_120 |
0.1988051103 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 17 |
Hb_000923_090 |
0.2019504023 |
- |
- |
- |
| 18 |
Hb_001146_010 |
0.2055286153 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 19 |
Hb_009898_040 |
0.2067641039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002344_130 |
0.2072891865 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |