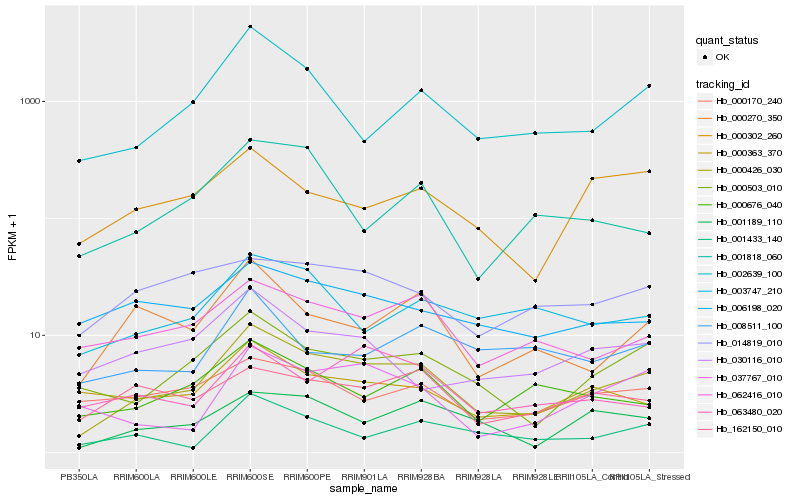
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000426_030 |
0.0 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000503_010 |
0.1506764132 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 3 |
Hb_002639_100 |
0.1608365552 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 4 |
Hb_062416_010 |
0.169676707 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 5 |
Hb_001189_110 |
0.1721067303 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 6 |
Hb_000270_350 |
0.1736492487 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 7 |
Hb_000676_040 |
0.1814437901 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 8 |
Hb_000363_370 |
0.1832633535 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 9 |
Hb_037767_010 |
0.1845838792 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001433_140 |
0.1849547012 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 11 |
Hb_003747_210 |
0.185195706 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 12 |
Hb_162150_010 |
0.1861434803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000170_240 |
0.1862490717 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 14 |
Hb_000302_260 |
0.1862886208 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 15 |
Hb_006198_020 |
0.1905113345 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 16 |
Hb_014819_010 |
0.1922351848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001818_060 |
0.1933865707 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 18 |
Hb_063480_020 |
0.197091656 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_030116_010 |
0.1981253841 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 20 |
Hb_008511_100 |
0.1983832618 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |