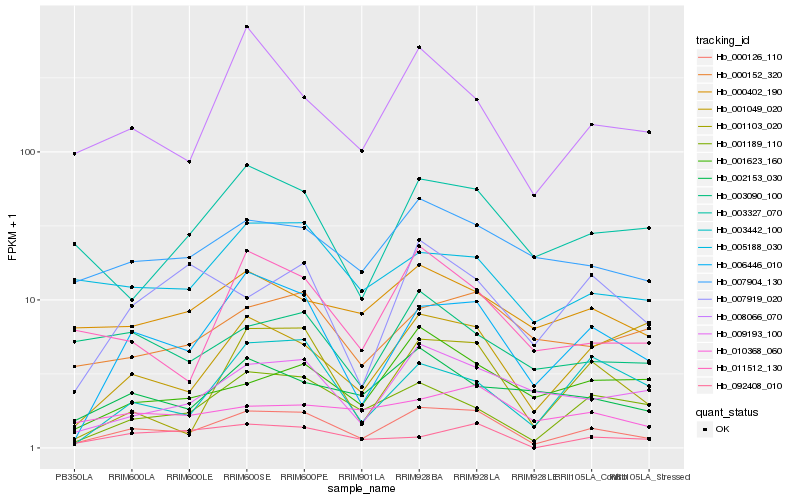
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000126_110 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 2 |
Hb_006446_010 |
0.1383460767 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_007904_130 |
0.1831316563 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 4 |
Hb_001103_020 |
0.1839859955 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 5 |
Hb_009193_100 |
0.1849784188 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 6 |
Hb_001189_110 |
0.1850215771 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 7 |
Hb_011512_130 |
0.1906877003 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 8 |
Hb_005188_030 |
0.1909440655 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_008066_070 |
0.1910358217 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 10 |
Hb_003327_070 |
0.193344486 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 11 |
Hb_000152_320 |
0.1963663096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000402_190 |
0.1999798508 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 13 |
Hb_007919_020 |
0.2013879842 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 14 |
Hb_092408_010 |
0.2020252799 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001049_020 |
0.2023088085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003090_100 |
0.2048571366 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 17 |
Hb_001623_160 |
0.2050522189 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 18 |
Hb_002153_030 |
0.2054877448 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 19 |
Hb_003442_100 |
0.206670935 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 20 |
Hb_010368_060 |
0.2102823815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |