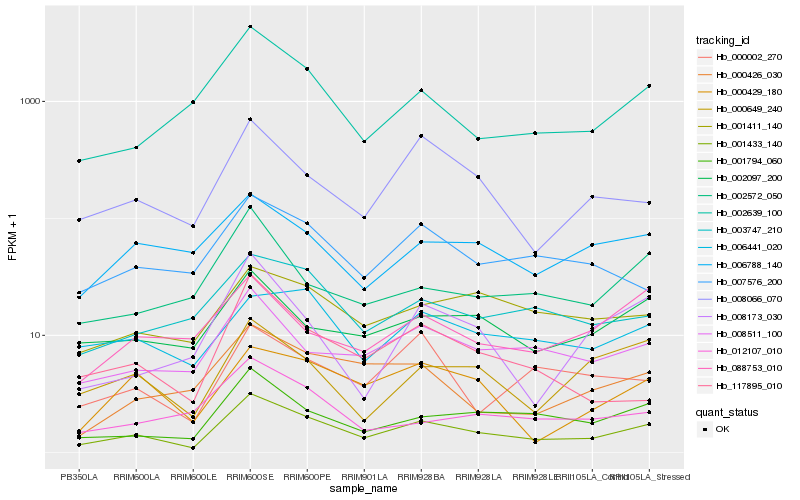
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_140 |
0.0 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 2 |
Hb_001794_060 |
0.1473145364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008511_100 |
0.1592363723 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 4 |
Hb_002639_100 |
0.1654251144 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 5 |
Hb_008066_070 |
0.1695725554 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 6 |
Hb_003747_210 |
0.1718690163 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 7 |
Hb_002097_200 |
0.1735597455 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_002572_050 |
0.1771647646 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_001411_140 |
0.1774739487 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 10 |
Hb_007576_200 |
0.1778436657 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000649_240 |
0.1779900173 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 12 |
Hb_000429_180 |
0.1801544457 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006441_020 |
0.1801967356 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 14 |
Hb_117895_010 |
0.1834478474 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 15 |
Hb_006788_140 |
0.1837731039 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 16 |
Hb_000426_030 |
0.1849547012 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000002_270 |
0.1890897974 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 18 |
Hb_012107_010 |
0.1895424244 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 19 |
Hb_008173_030 |
0.1905049736 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 20 |
Hb_088753_010 |
0.1933212158 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |