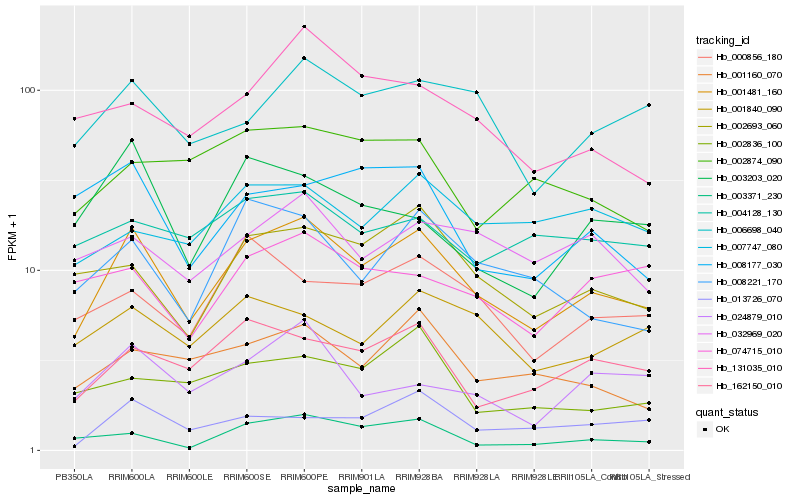
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001481_160 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 2 |
Hb_162150_010 |
0.1408176792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002693_060 |
0.1409646607 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_006698_040 |
0.1464180785 |
- |
- |
bHLH1 transcription factor [Hevea brasiliensis] |
| 5 |
Hb_001160_070 |
0.148144463 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 6 |
Hb_008221_170 |
0.1484310513 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 7 |
Hb_000856_180 |
0.1579944535 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 8 |
Hb_131035_010 |
0.1595678859 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001840_090 |
0.1604644619 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_013726_070 |
0.1615187101 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Jatropha curcas] |
| 11 |
Hb_032969_020 |
0.1622871985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003203_020 |
0.1629137373 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_003371_230 |
0.1640849904 |
- |
- |
lactase-like b precursor [Scleropages formosus] |
| 14 |
Hb_024879_010 |
0.1663028535 |
- |
- |
unknown [Glycine max] |
| 15 |
Hb_002874_090 |
0.1670281519 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 16 |
Hb_008177_030 |
0.1671679868 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_074715_010 |
0.1684055996 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004128_130 |
0.1686905337 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002836_100 |
0.1693925886 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007747_080 |
0.1696950773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |