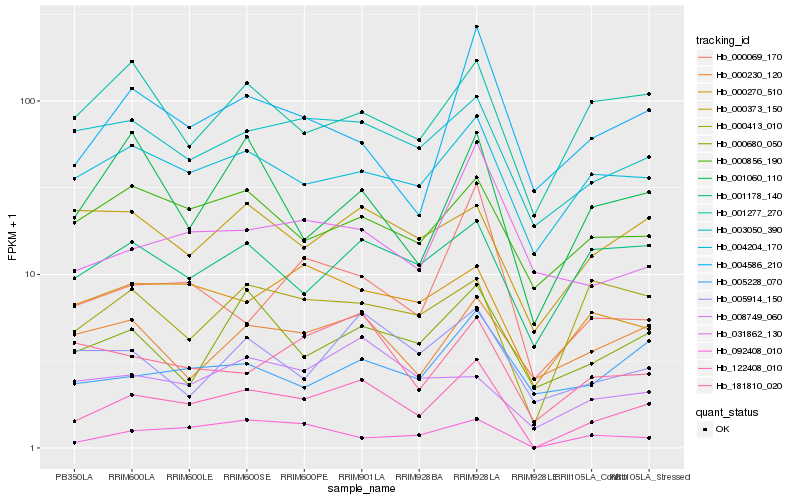
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122408_010 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004204_170 |
0.1817672565 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 3 |
Hb_003050_390 |
0.1822744187 |
- |
- |
PREDICTED: uncharacterized protein LOC104442543 [Eucalyptus grandis] |
| 4 |
Hb_181810_020 |
0.1865135847 |
- |
- |
- |
| 5 |
Hb_092408_010 |
0.1905047459 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001178_140 |
0.1942407018 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000680_050 |
0.1964242197 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 8 |
Hb_000856_190 |
0.1967551814 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 9 |
Hb_005914_150 |
0.1971782162 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 10 |
Hb_000270_510 |
0.1975005352 |
- |
- |
hypothetical protein CISIN_1g008173mg [Citrus sinensis] |
| 11 |
Hb_000230_120 |
0.1993785247 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 12 |
Hb_004586_210 |
0.2005555399 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000413_010 |
0.2010384386 |
- |
- |
hypothetical protein JCGZ_21316 [Jatropha curcas] |
| 14 |
Hb_031862_130 |
0.2019867422 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000373_150 |
0.202533065 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 16 |
Hb_000069_170 |
0.2035284392 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 17 |
Hb_008749_060 |
0.2049489715 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 18 |
Hb_001060_110 |
0.2052092718 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 19 |
Hb_005228_070 |
0.2059377138 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g13230, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001277_270 |
0.2059954584 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |