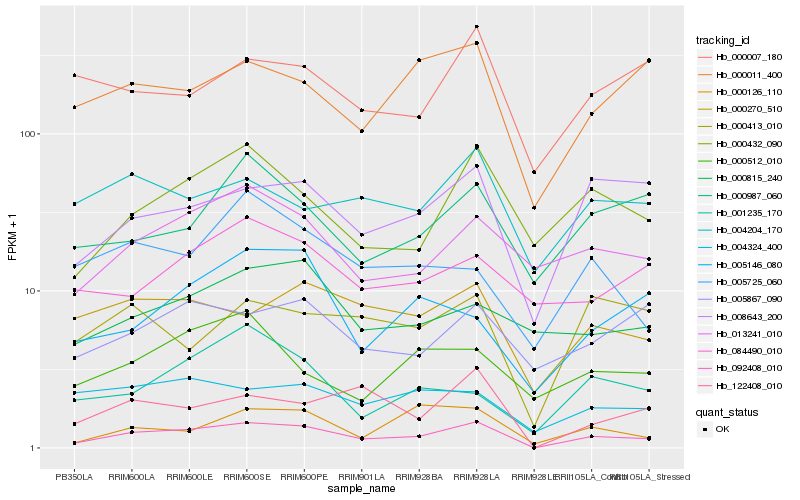
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092408_010 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_008643_200 |
0.17941283 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 [Vitis vinifera] |
| 3 |
Hb_000432_090 |
0.1856210982 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_013241_010 |
0.1896853262 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_122408_010 |
0.1905047459 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000270_510 |
0.1924764651 |
- |
- |
hypothetical protein CISIN_1g008173mg [Citrus sinensis] |
| 7 |
Hb_000011_400 |
0.1998630688 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 4 [Hevea brasiliensis] |
| 8 |
Hb_000126_110 |
0.2020252799 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 9 |
Hb_000413_010 |
0.2054461597 |
- |
- |
hypothetical protein JCGZ_21316 [Jatropha curcas] |
| 10 |
Hb_005867_090 |
0.207626177 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 11 |
Hb_000987_060 |
0.2077140702 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 12 |
Hb_004204_170 |
0.208974583 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 13 |
Hb_004324_400 |
0.209884196 |
- |
- |
PREDICTED: protein timeless homolog [Jatropha curcas] |
| 14 |
Hb_001235_170 |
0.2126277636 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 15 |
Hb_005146_080 |
0.2132386712 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000815_240 |
0.2133244462 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000512_010 |
0.2144754153 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 18 |
Hb_084490_010 |
0.2151784973 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005725_060 |
0.2152990602 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 20 |
Hb_000007_180 |
0.2153673491 |
- |
- |
hypothetical protein PRUPE_ppb024585mg, partial [Prunus persica] |