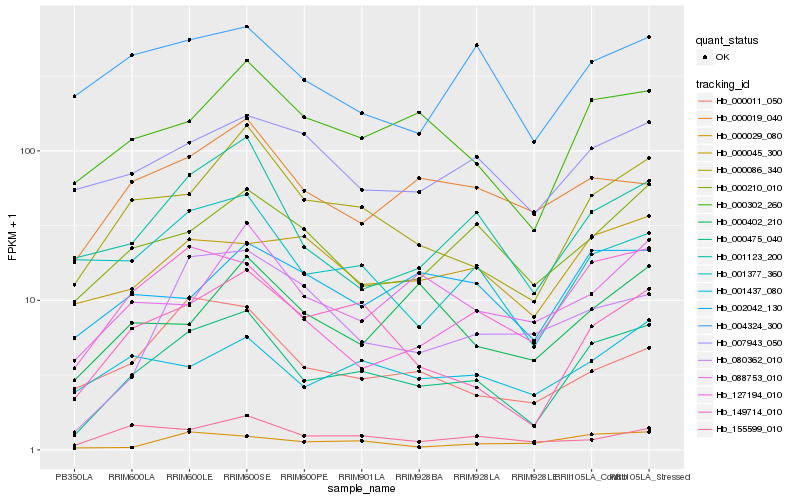
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000475_040 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000086_340 |
0.1385238391 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 3 |
Hb_149714_010 |
0.1533568808 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 4 |
Hb_000302_260 |
0.158620467 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 5 |
Hb_127194_010 |
0.1663858944 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 6 |
Hb_001123_200 |
0.1681243433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000210_010 |
0.1827429777 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 8 |
Hb_001377_360 |
0.1878654201 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 9 |
Hb_000029_080 |
0.1921246597 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 10 |
Hb_000019_040 |
0.1936797913 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 11 |
Hb_155599_010 |
0.1946551511 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000011_050 |
0.1952572439 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 13 |
Hb_007943_050 |
0.1952580999 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 14 |
Hb_001437_080 |
0.1956505099 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_05472 [Jatropha curcas] |
| 15 |
Hb_088753_010 |
0.1984692885 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 16 |
Hb_000045_300 |
0.2012514224 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 17 |
Hb_080362_010 |
0.201377611 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 18 |
Hb_004324_300 |
0.2022626033 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 19 |
Hb_002042_130 |
0.2024979282 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 20 |
Hb_000402_210 |
0.2029768047 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |