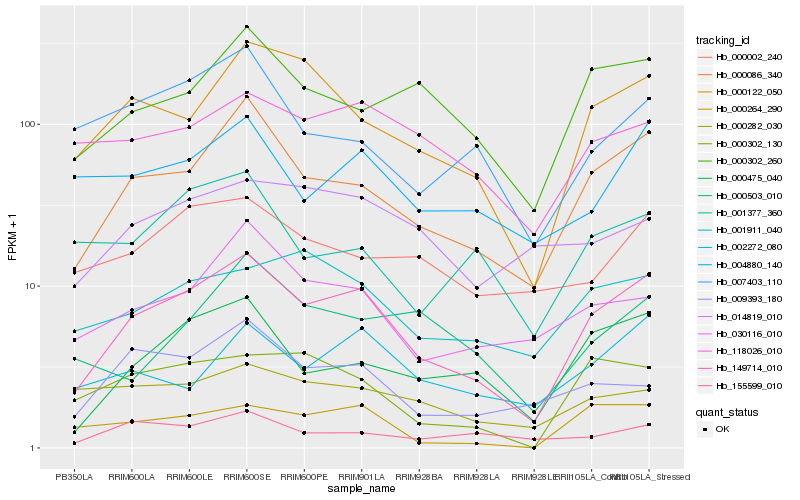
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149714_010 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 2 |
Hb_000086_340 |
0.1059725071 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 3 |
Hb_000475_040 |
0.1533568808 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000122_050 |
0.1545997277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000302_260 |
0.1712882301 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 6 |
Hb_030116_010 |
0.1759069456 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 7 |
Hb_000264_290 |
0.1803194403 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 8 |
Hb_000302_130 |
0.1820864558 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_002272_080 |
0.1838282917 |
- |
- |
- |
| 10 |
Hb_001911_040 |
0.1841201112 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 11 |
Hb_009393_180 |
0.1869364111 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_004880_140 |
0.1882968478 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 13 |
Hb_118026_010 |
0.1930458405 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 14 |
Hb_000002_240 |
0.1936246443 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 15 |
Hb_155599_010 |
0.1943935082 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000503_010 |
0.1959886137 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 17 |
Hb_007403_110 |
0.1974800687 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 18 |
Hb_014819_010 |
0.2020068889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_360 |
0.2022334391 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 20 |
Hb_000282_030 |
0.2044476499 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |