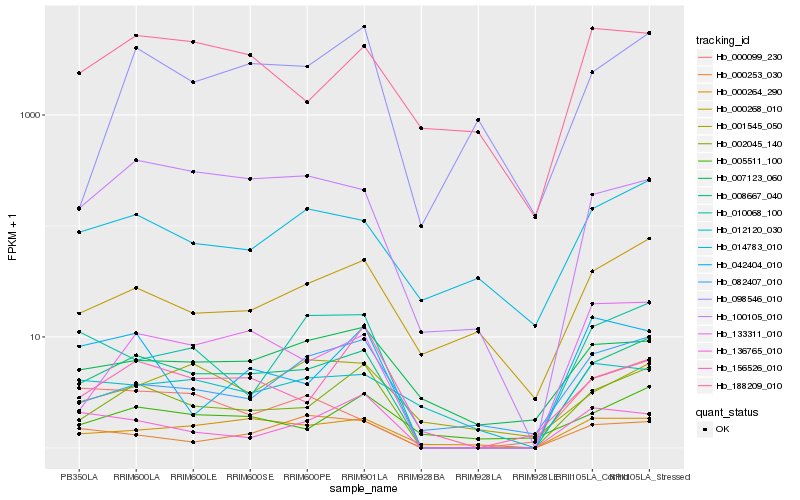
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008667_040 |
0.0 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 2 |
Hb_002045_140 |
0.1189494069 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 3 |
Hb_000264_290 |
0.1750674674 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 4 |
Hb_100105_010 |
0.1767857322 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 5 |
Hb_133311_010 |
0.1937813447 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 6 |
Hb_010068_100 |
0.2024215399 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007123_060 |
0.2027140034 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 8 |
Hb_014783_010 |
0.2062670312 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000268_010 |
0.2081749806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_082407_010 |
0.210051211 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_000253_030 |
0.2119574121 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 12 |
Hb_005511_100 |
0.2139167536 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 13 |
Hb_188209_010 |
0.2141328799 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_156526_010 |
0.2144734906 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 15 |
Hb_136765_010 |
0.2198212352 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_012120_030 |
0.2235523962 |
- |
- |
- |
| 17 |
Hb_098546_010 |
0.2255751894 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 18 |
Hb_000099_230 |
0.2259439924 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 19 |
Hb_001545_050 |
0.2267384668 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 20 |
Hb_042404_010 |
0.2268151037 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |