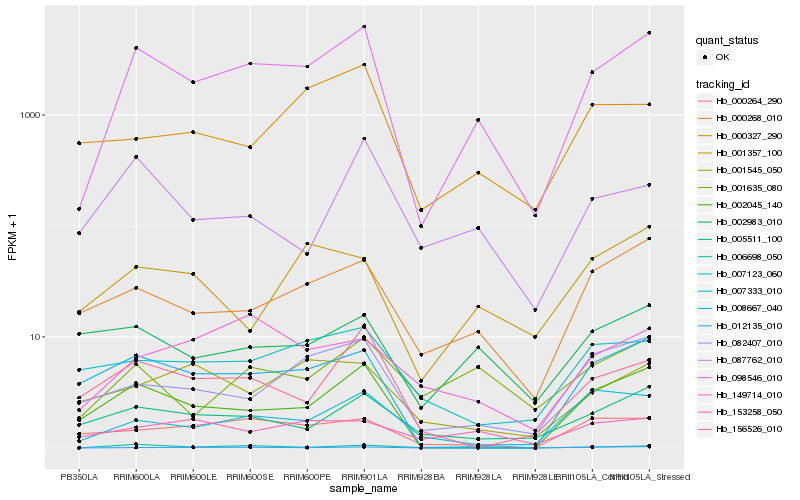
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098546_010 |
0.0 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 2 |
Hb_002045_140 |
0.203076666 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 3 |
Hb_000268_010 |
0.2060551994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_082407_010 |
0.2119765731 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 5 |
Hb_005511_100 |
0.2129054634 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006698_050 |
0.221193289 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 7 |
Hb_007333_010 |
0.2255135078 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_008667_040 |
0.2255751894 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 9 |
Hb_001635_080 |
0.2284208371 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.1-like [Jatropha curcas] |
| 10 |
Hb_012135_010 |
0.2310399802 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 11 |
Hb_156526_010 |
0.2316136485 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 12 |
Hb_149714_010 |
0.2347369514 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 13 |
Hb_000264_290 |
0.2367332612 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 14 |
Hb_007123_060 |
0.2368031632 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 15 |
Hb_002983_010 |
0.2446777902 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 16 |
Hb_087762_010 |
0.2451145859 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 17 |
Hb_000327_290 |
0.2463728426 |
- |
- |
Histone H4 [Triticum urartu] |
| 18 |
Hb_001545_050 |
0.2468364242 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 19 |
Hb_001357_100 |
0.2472531048 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 20 |
Hb_153258_050 |
0.2489845517 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |