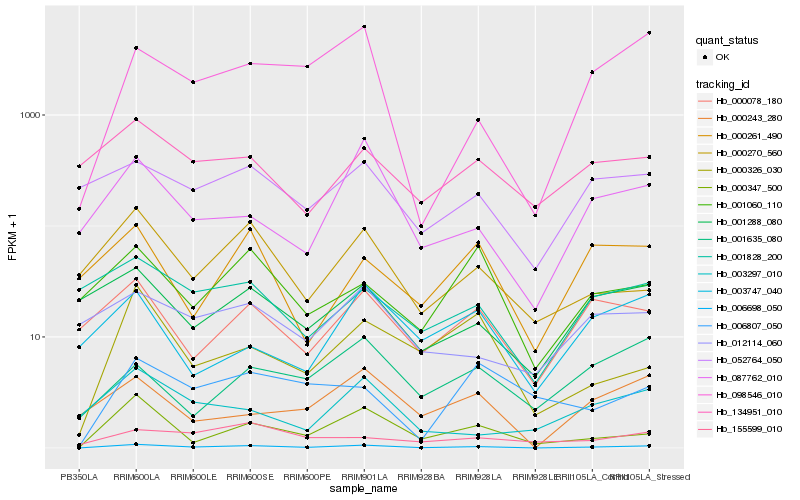
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006698_050 |
0.0 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_000347_500 |
0.2063524561 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_098546_010 |
0.221193289 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 4 |
Hb_087762_010 |
0.2489104893 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 5 |
Hb_000243_280 |
0.2511271004 |
- |
- |
- |
| 6 |
Hb_000078_180 |
0.2532487576 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 7 |
Hb_000326_030 |
0.254641402 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |
| 8 |
Hb_000270_560 |
0.2546591447 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 9 |
Hb_052764_050 |
0.2667533616 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 10 |
Hb_001288_080 |
0.267468501 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 11 |
Hb_001635_080 |
0.2687102726 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.1-like [Jatropha curcas] |
| 12 |
Hb_001060_110 |
0.2715177099 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 13 |
Hb_000261_490 |
0.2742696376 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 14 |
Hb_003297_010 |
0.2771963759 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 15 |
Hb_155599_010 |
0.2790247608 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_001828_200 |
0.2792675485 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 17 |
Hb_003747_040 |
0.2800460257 |
- |
- |
- |
| 18 |
Hb_012114_060 |
0.280118512 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 19 |
Hb_006807_050 |
0.280981052 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPH8A, putative [Ricinus communis] |
| 20 |
Hb_134951_010 |
0.2825393554 |
- |
- |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |