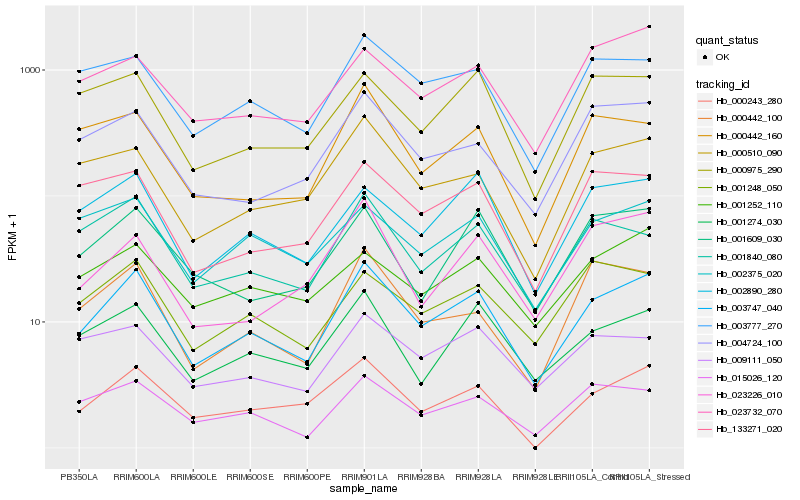
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003747_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_015026_120 |
0.1159311695 |
- |
- |
hypothetical protein TRIUR3_03502 [Triticum urartu] |
| 3 |
Hb_001274_030 |
0.1169451728 |
- |
- |
PREDICTED: uncharacterized protein LOC105628035 [Jatropha curcas] |
| 4 |
Hb_003777_270 |
0.1181667839 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 5 |
Hb_002890_280 |
0.1217917761 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 6 |
Hb_000243_280 |
0.1229054994 |
- |
- |
- |
| 7 |
Hb_000510_090 |
0.1236492362 |
- |
- |
40S ribosomal protein S15 [Morus notabilis] |
| 8 |
Hb_004724_100 |
0.1270730661 |
- |
- |
60S ribosomal protein L10aA [Hevea brasiliensis] |
| 9 |
Hb_001840_080 |
0.128273928 |
- |
- |
hypothetical protein POPTR_0008s14370g [Populus trichocarpa] |
| 10 |
Hb_000442_100 |
0.1284766378 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 11 |
Hb_001609_030 |
0.1308220459 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001248_050 |
0.1316120066 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 13 |
Hb_133271_020 |
0.131668508 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 14 |
Hb_023732_070 |
0.1322520527 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 15 |
Hb_002375_020 |
0.1324632974 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 16 |
Hb_000442_160 |
0.1331174317 |
- |
- |
alpha-soluble nsf attachment protein, putative [Ricinus communis] |
| 17 |
Hb_000975_290 |
0.1331997423 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 18 |
Hb_001252_110 |
0.1341003258 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_009111_050 |
0.1343758164 |
transcription factor |
TF Family: Jumonji |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540 [Populus euphratica] |
| 20 |
Hb_023226_010 |
0.1359701989 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-7-like isoform X1 [Jatropha curcas] |