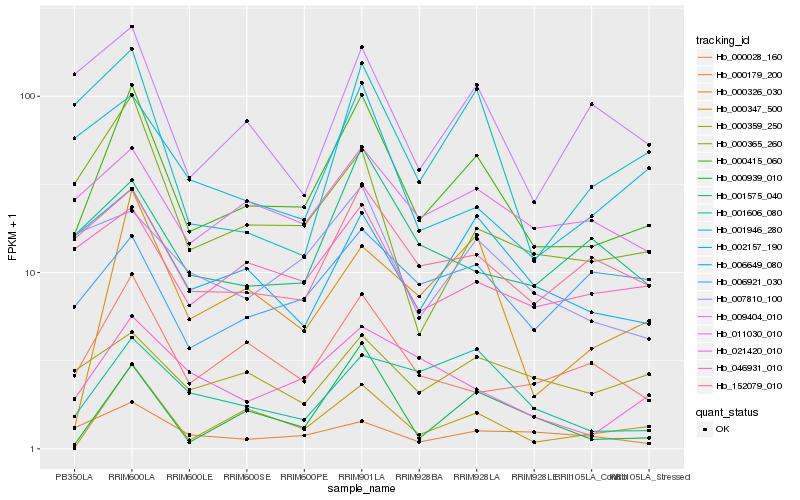
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000415_060 |
0.0 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 2 |
Hb_000028_160 |
0.16807568 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 3 |
Hb_152079_010 |
0.1718582238 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 4 |
Hb_000347_500 |
0.1767107416 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011030_010 |
0.1817113385 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_046931_010 |
0.1828466877 |
- |
- |
hypothetical protein JCGZ_26924 [Jatropha curcas] |
| 7 |
Hb_021420_010 |
0.187922881 |
- |
- |
hypothetical protein CICLE_v10029508mg [Citrus clementina] |
| 8 |
Hb_006649_080 |
0.1882537505 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 9 |
Hb_000365_260 |
0.1885025839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000326_030 |
0.1908989659 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |
| 11 |
Hb_001575_040 |
0.1937011051 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007810_100 |
0.195214708 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 13 |
Hb_000359_250 |
0.198388455 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g26790, mitochondrial [Vitis vinifera] |
| 14 |
Hb_009404_010 |
0.1986177536 |
- |
- |
PREDICTED: casein kinase II subunit alpha-2-like [Glycine max] |
| 15 |
Hb_002157_190 |
0.1992569184 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 16 |
Hb_001606_080 |
0.2019711911 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000179_200 |
0.2024316903 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000939_010 |
0.2030515633 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 19 |
Hb_001946_280 |
0.2032800211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006921_030 |
0.2034207122 |
- |
- |
PREDICTED: uncharacterized protein LOC105638271 [Jatropha curcas] |