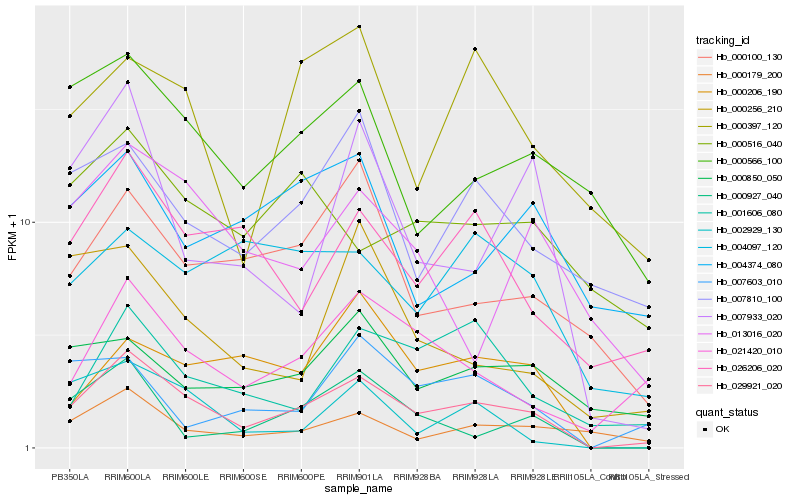
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029921_020 |
0.0 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 2 |
Hb_026206_020 |
0.1973994562 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X1 [Jatropha curcas] |
| 3 |
Hb_021420_010 |
0.197883404 |
- |
- |
hypothetical protein CICLE_v10029508mg [Citrus clementina] |
| 4 |
Hb_007810_100 |
0.2109343781 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 5 |
Hb_000179_200 |
0.2119741263 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000566_100 |
0.2125634829 |
- |
- |
hypothetical protein JCGZ_17993 [Jatropha curcas] |
| 7 |
Hb_000100_130 |
0.2158942544 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 8 |
Hb_002929_130 |
0.2206326871 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 9 |
Hb_001606_080 |
0.2211192664 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000397_120 |
0.2223961652 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 11 |
Hb_000850_050 |
0.223561906 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 12 |
Hb_000206_190 |
0.2236421782 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 13 |
Hb_013016_020 |
0.2294937307 |
- |
- |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 14 |
Hb_000927_040 |
0.2295407271 |
- |
- |
- |
| 15 |
Hb_007933_020 |
0.2296706769 |
- |
- |
PREDICTED: uncharacterized protein At4g37920, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004097_120 |
0.2309266344 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_007603_010 |
0.2344373196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000516_040 |
0.2361146161 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000256_210 |
0.2364208589 |
- |
- |
PREDICTED: 40S ribosomal protein S11-like [Phoenix dactylifera] |
| 20 |
Hb_004374_080 |
0.2365818242 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |