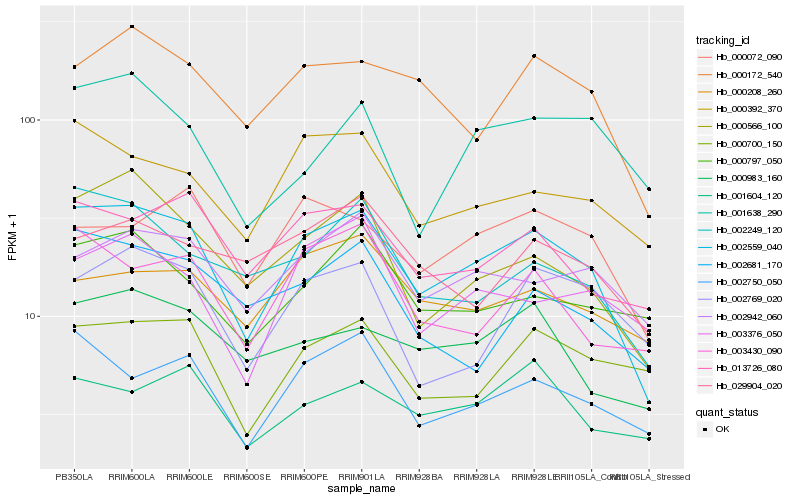
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002559_040 |
0.0 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002750_050 |
0.1202247846 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 3 |
Hb_000566_100 |
0.1204222921 |
- |
- |
hypothetical protein JCGZ_17993 [Jatropha curcas] |
| 4 |
Hb_000172_540 |
0.1233580265 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 5 |
Hb_013726_080 |
0.1247076013 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 6 |
Hb_000392_370 |
0.1271809242 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 7 |
Hb_002249_120 |
0.132555831 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 8 |
Hb_002942_060 |
0.1326374498 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 9 |
Hb_003430_090 |
0.1333512757 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000072_090 |
0.1335481679 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 11 |
Hb_000983_160 |
0.1338244366 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 12 |
Hb_003376_050 |
0.1410985353 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 13 |
Hb_001604_120 |
0.1426761411 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000797_050 |
0.1434177254 |
- |
- |
PREDICTED: uncharacterized protein LOC105645916 [Jatropha curcas] |
| 15 |
Hb_002681_170 |
0.143997456 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 16 |
Hb_000700_150 |
0.1444702362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002769_020 |
0.1458651798 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 18 |
Hb_000208_260 |
0.1482174308 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_001638_290 |
0.148384607 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_029904_020 |
0.151179149 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |