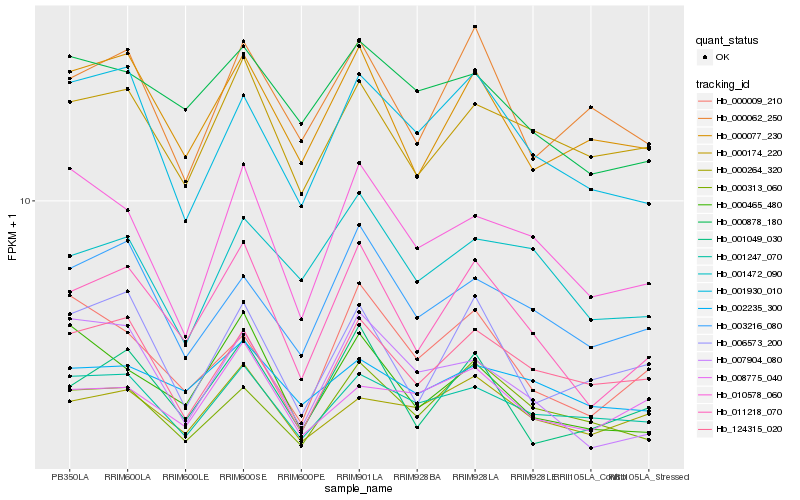
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 2 |
Hb_001049_030 |
0.104429201 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_000077_230 |
0.1158479767 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 4 |
Hb_003216_080 |
0.1201910238 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560-like [Jatropha curcas] |
| 5 |
Hb_000465_480 |
0.1209972449 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001930_010 |
0.1215262395 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 7 |
Hb_000264_320 |
0.1243351467 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 8 |
Hb_000313_060 |
0.1253693119 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_124315_020 |
0.1256225663 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001247_070 |
0.1264464984 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g56690, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000174_220 |
0.1267156412 |
- |
- |
cohesin subunit rad21, putative [Ricinus communis] |
| 12 |
Hb_006573_200 |
0.1272786849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 13 |
Hb_010578_060 |
0.1277633284 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 14 |
Hb_000009_210 |
0.1298473122 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01390 [Jatropha curcas] |
| 15 |
Hb_002235_300 |
0.1302632582 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007904_080 |
0.131740763 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 17 |
Hb_000878_180 |
0.1318174681 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001472_090 |
0.1339391631 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008775_040 |
0.1356388917 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 20 |
Hb_000062_250 |
0.1358443251 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |