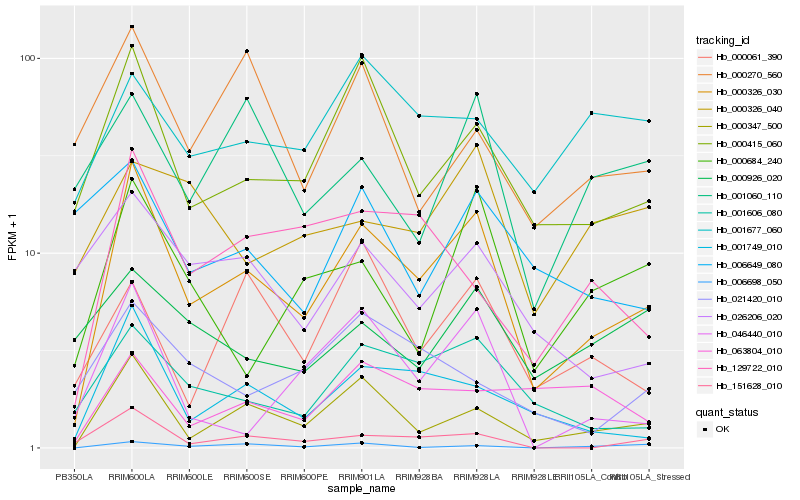
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000326_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |
| 2 |
Hb_000347_500 |
0.1602332102 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000415_060 |
0.1908989659 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 4 |
Hb_001749_010 |
0.1959799486 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 5 |
Hb_001606_080 |
0.2081488225 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 6 |
Hb_151628_010 |
0.2175598194 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_129722_010 |
0.2325553141 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 8 |
Hb_000684_240 |
0.2373746864 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 9 |
Hb_046440_010 |
0.2410087588 |
- |
- |
hypothetical protein JCGZ_06190 [Jatropha curcas] |
| 10 |
Hb_026206_020 |
0.249657997 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006698_050 |
0.254641402 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 12 |
Hb_000270_560 |
0.261779617 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 13 |
Hb_021420_010 |
0.2620621043 |
- |
- |
hypothetical protein CICLE_v10029508mg [Citrus clementina] |
| 14 |
Hb_001677_060 |
0.2645862418 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 15 |
Hb_001060_110 |
0.2650125835 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 16 |
Hb_000061_390 |
0.2652346038 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 17 |
Hb_063804_010 |
0.2672119252 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_000926_020 |
0.2702709916 |
- |
- |
- |
| 19 |
Hb_000326_040 |
0.2716731266 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 20 |
Hb_006649_080 |
0.2742664987 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |