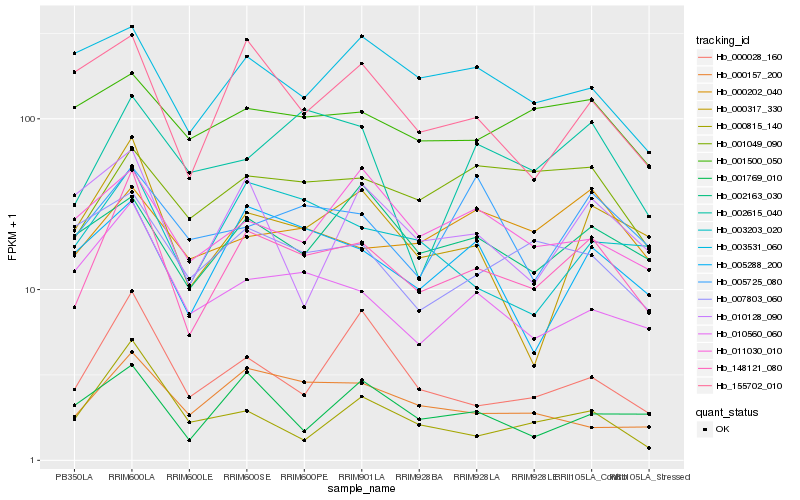
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148121_080 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 2 |
Hb_000317_330 |
0.1529385312 |
- |
- |
PREDICTED: RNA-binding protein 38-like [Jatropha curcas] |
| 3 |
Hb_010560_060 |
0.1650296685 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 4 |
Hb_000157_200 |
0.1716242791 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 5 |
Hb_005288_200 |
0.1726018305 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_003203_020 |
0.1753806963 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_000028_160 |
0.1777083668 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 8 |
Hb_155702_010 |
0.1814638066 |
- |
- |
Tetraspanin-5 [Gossypium arboreum] |
| 9 |
Hb_001049_090 |
0.1830825557 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_007803_060 |
0.1869166621 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 11 |
Hb_000815_140 |
0.1874782461 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_002163_030 |
0.1886642948 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 59 kDa regulatory subunit B' gamma isoform [Jatropha curcas] |
| 13 |
Hb_001769_010 |
0.1888718852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002615_040 |
0.1889800726 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 15 |
Hb_001500_050 |
0.1919014799 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 16 |
Hb_010128_090 |
0.1931424453 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 17 |
Hb_011030_010 |
0.1933966425 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000202_040 |
0.1945130573 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 19 |
Hb_003531_060 |
0.1964308097 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_005725_080 |
0.1964789751 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |