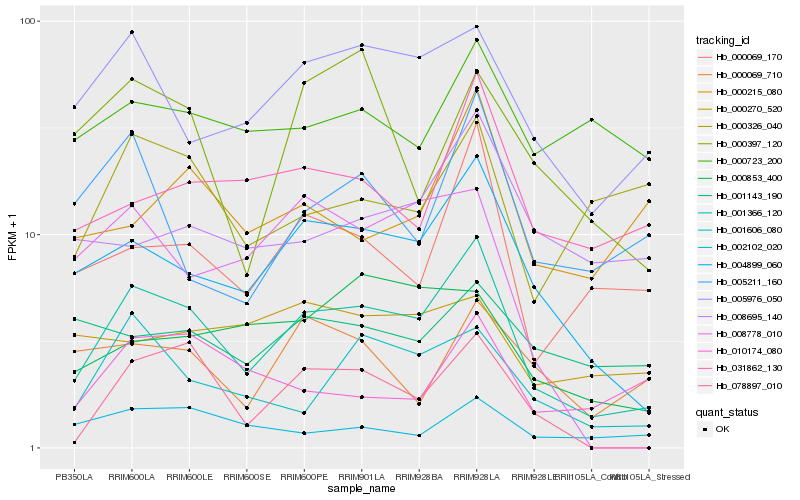
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_120 |
0.0 |
- |
- |
unknown [Zea mays] |
| 2 |
Hb_004899_060 |
0.1402821663 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 3 |
Hb_005976_050 |
0.1852923222 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 4 |
Hb_000069_170 |
0.1898156686 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 5 |
Hb_001606_080 |
0.1938748291 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 6 |
Hb_078897_010 |
0.198649171 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 7 |
Hb_031862_130 |
0.2060460614 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002102_020 |
0.2172242005 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 9 |
Hb_005211_160 |
0.2174562231 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 10 |
Hb_000853_400 |
0.2217657132 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 11 |
Hb_000397_120 |
0.2226193658 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 12 |
Hb_000215_080 |
0.2243265884 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008778_010 |
0.2243336509 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_008695_140 |
0.224809122 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 15 |
Hb_000069_710 |
0.2285135235 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 16 |
Hb_000326_040 |
0.2306490251 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 17 |
Hb_010174_080 |
0.2322281318 |
- |
- |
- |
| 18 |
Hb_000723_200 |
0.2334296922 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001143_190 |
0.2337794901 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 20 |
Hb_000270_520 |
0.2362275675 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |