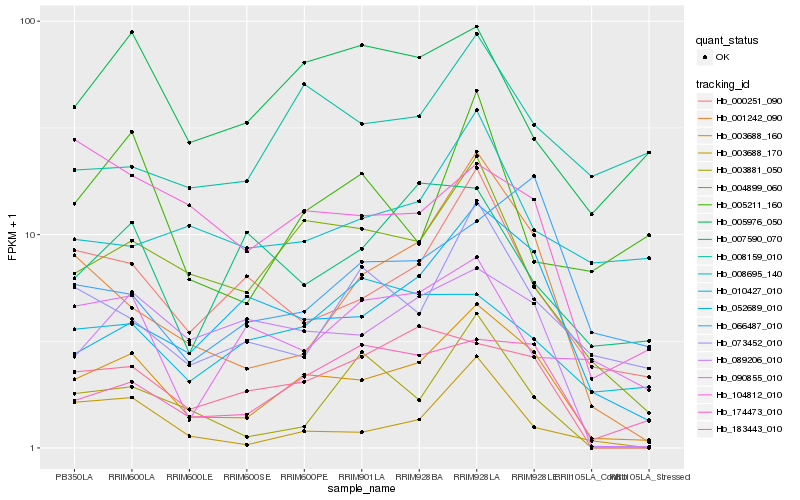
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_160 |
0.0 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 2 |
Hb_004899_060 |
0.170855103 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 3 |
Hb_052689_010 |
0.1795882799 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 4 |
Hb_174473_010 |
0.1804219827 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 5 |
Hb_001242_090 |
0.1835466691 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 6 |
Hb_000251_090 |
0.1839997899 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 7 |
Hb_089206_010 |
0.1877081838 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_183443_010 |
0.200639632 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 9 |
Hb_073452_010 |
0.2145362769 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_003688_170 |
0.218210534 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 11 |
Hb_005976_050 |
0.2259411116 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 12 |
Hb_104812_010 |
0.2271983743 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Populus euphratica] |
| 13 |
Hb_090855_010 |
0.2377277091 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_003881_050 |
0.2380572225 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 15 |
Hb_007590_070 |
0.2405344453 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 16 |
Hb_010427_010 |
0.2410293884 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 17 |
Hb_066487_010 |
0.2415054304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005211_160 |
0.241841939 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 19 |
Hb_008695_140 |
0.2421429279 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 20 |
Hb_008159_010 |
0.2422154069 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |