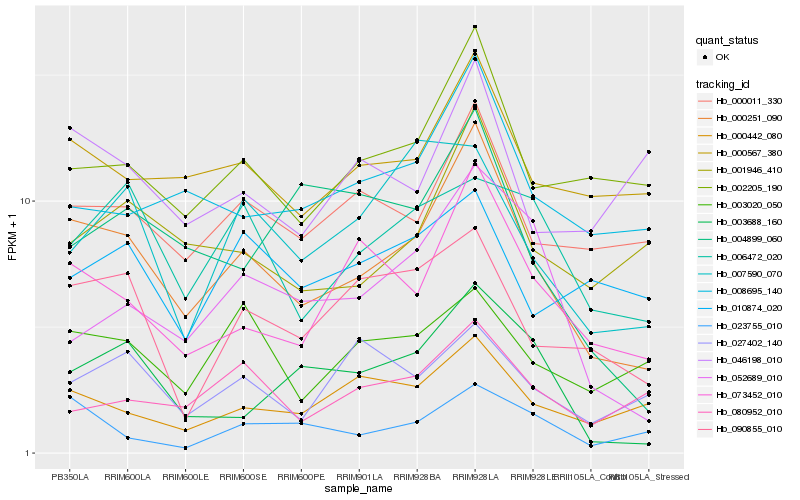
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_090 |
0.0 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 2 |
Hb_073452_010 |
0.1478785552 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_002205_190 |
0.1587837152 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 4 |
Hb_000567_380 |
0.1604118474 |
- |
- |
ubiquitin specific protease 39 and snrnp assembly factor, putative [Ricinus communis] |
| 5 |
Hb_000011_330 |
0.1620629462 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 6 |
Hb_001946_410 |
0.1650042003 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008695_140 |
0.1688778424 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 8 |
Hb_090855_010 |
0.1724845847 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_000442_080 |
0.1824703282 |
- |
- |
hypothetical protein JCGZ_03373 [Jatropha curcas] |
| 10 |
Hb_003688_160 |
0.1839997899 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 11 |
Hb_052689_010 |
0.1876644721 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 12 |
Hb_003020_050 |
0.1895596356 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_004899_060 |
0.1918782328 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 14 |
Hb_006472_020 |
0.1936678794 |
- |
- |
heat shock protein [Cicer arietinum] |
| 15 |
Hb_007590_070 |
0.1954898924 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 16 |
Hb_046198_010 |
0.1955712992 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 17 |
Hb_080952_010 |
0.1971869446 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20770 [Jatropha curcas] |
| 18 |
Hb_023755_010 |
0.2018523062 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_027402_140 |
0.2025466481 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g47530 [Jatropha curcas] |
| 20 |
Hb_010874_020 |
0.2043992714 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |