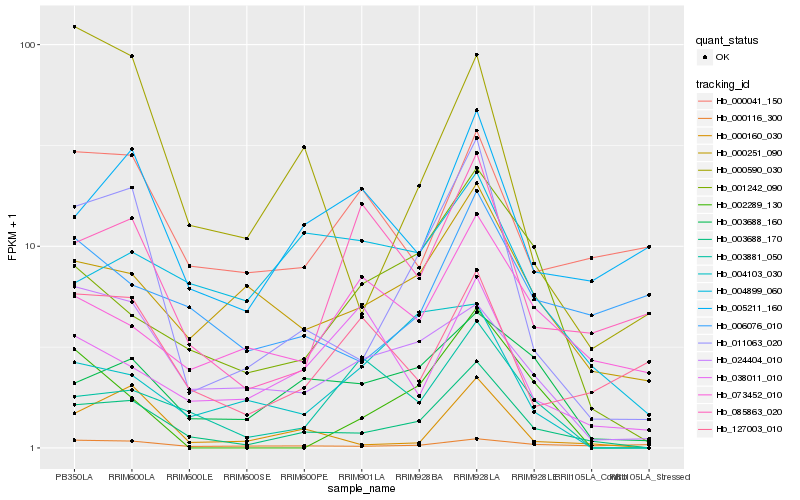
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_170 |
0.0 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 2 |
Hb_011063_020 |
0.1528097837 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_003688_160 |
0.218210534 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 4 |
Hb_000251_090 |
0.2245471657 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 5 |
Hb_001242_090 |
0.2269946033 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 6 |
Hb_000160_030 |
0.2277478066 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 7 |
Hb_024404_010 |
0.2378784447 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 8 |
Hb_005211_160 |
0.2548095722 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 9 |
Hb_003881_050 |
0.2582250693 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 10 |
Hb_085863_020 |
0.259740278 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 11 |
Hb_073452_010 |
0.272847846 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000590_030 |
0.2730220524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004899_060 |
0.2730812373 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 14 |
Hb_127003_010 |
0.2742774908 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 15 |
Hb_006076_010 |
0.274349424 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_000116_300 |
0.2746291775 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000041_150 |
0.2769506972 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 18 |
Hb_002289_130 |
0.2797794114 |
transcription factor |
TF Family: C2H2 |
arsenite inducuble RNA associated protein aip-1, putative [Ricinus communis] |
| 19 |
Hb_004103_030 |
0.2887509603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_038011_010 |
0.2891000084 |
- |
- |
hypothetical protein POPTR_0031s004401g, partial [Populus trichocarpa] |