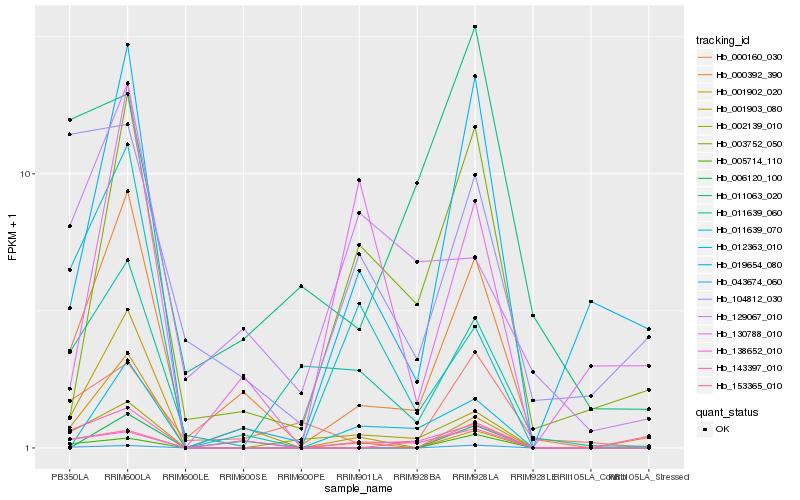
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_390 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001903_080 |
0.2496979716 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 3 |
Hb_019654_080 |
0.2670766883 |
- |
- |
- |
| 4 |
Hb_003752_050 |
0.2762442068 |
- |
- |
hypothetical protein JCGZ_15078 [Jatropha curcas] |
| 5 |
Hb_002139_010 |
0.2842145531 |
- |
- |
- |
| 6 |
Hb_043674_060 |
0.2842996425 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 7 |
Hb_000160_030 |
0.2859076058 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 8 |
Hb_011639_070 |
0.2939496212 |
- |
- |
PREDICTED: uncharacterized protein LOC105636024 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006120_100 |
0.2988917229 |
- |
- |
- |
| 10 |
Hb_011639_060 |
0.3048880182 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 11 |
Hb_129067_010 |
0.3072761025 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 12 |
Hb_138652_010 |
0.3095745583 |
- |
- |
hypothetical protein JCGZ_13474 [Jatropha curcas] |
| 13 |
Hb_005714_110 |
0.31057893 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA10 [Jatropha curcas] |
| 14 |
Hb_012363_010 |
0.3156292555 |
- |
- |
- |
| 15 |
Hb_153365_010 |
0.320627814 |
- |
- |
PREDICTED: uncharacterized protein LOC104236681 [Nicotiana sylvestris] |
| 16 |
Hb_143397_010 |
0.3267248806 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 17 |
Hb_011063_020 |
0.3309235295 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 18 |
Hb_104812_030 |
0.3330890338 |
- |
- |
hypothetical protein POPTR_0178s00210g [Populus trichocarpa] |
| 19 |
Hb_001902_020 |
0.3347026392 |
- |
- |
- |
| 20 |
Hb_130788_010 |
0.3458757813 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |