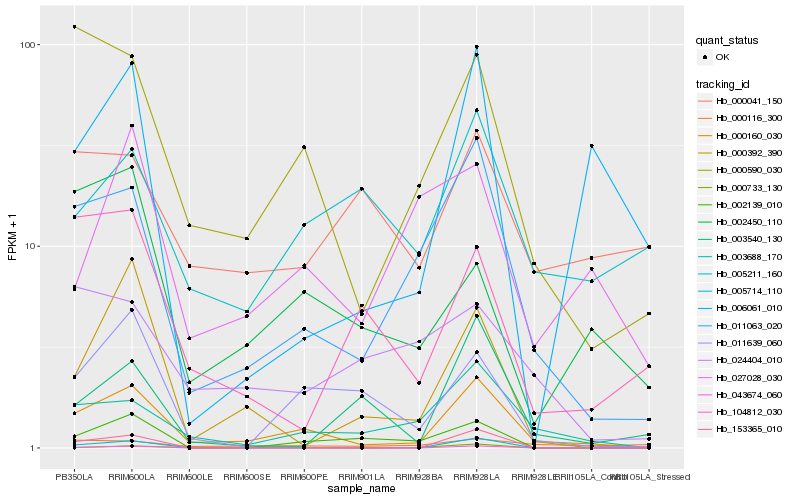
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_030 |
0.0 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 2 |
Hb_011063_020 |
0.2007788297 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_003688_170 |
0.2277478066 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 4 |
Hb_000590_030 |
0.2314768824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011639_060 |
0.259744755 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 6 |
Hb_000392_390 |
0.2859076058 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_002139_010 |
0.2915667563 |
- |
- |
- |
| 8 |
Hb_006061_010 |
0.3093470995 |
- |
- |
PREDICTED: phytosulfokines 4-like [Populus euphratica] |
| 9 |
Hb_002450_110 |
0.3101505463 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_027028_030 |
0.3120656503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005714_110 |
0.3138338693 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA10 [Jatropha curcas] |
| 12 |
Hb_153365_010 |
0.3139422039 |
- |
- |
PREDICTED: uncharacterized protein LOC104236681 [Nicotiana sylvestris] |
| 13 |
Hb_005211_160 |
0.3140567214 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 14 |
Hb_043674_060 |
0.3152223595 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 15 |
Hb_003540_130 |
0.3170801422 |
- |
- |
- |
| 16 |
Hb_000733_130 |
0.3200622909 |
- |
- |
PREDICTED: cucumisin [Vitis vinifera] |
| 17 |
Hb_024404_010 |
0.325332291 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 18 |
Hb_104812_030 |
0.3264970133 |
- |
- |
hypothetical protein POPTR_0178s00210g [Populus trichocarpa] |
| 19 |
Hb_000116_300 |
0.3345546936 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000041_150 |
0.3350866169 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |