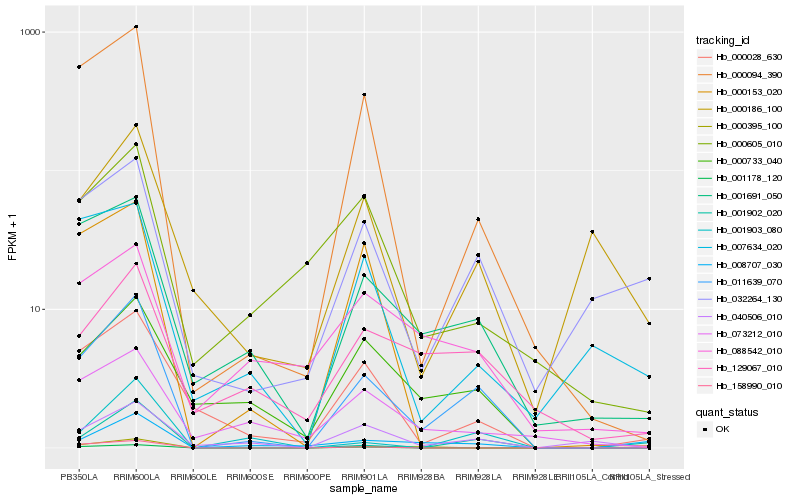
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011639_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636024 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000094_390 |
0.1928924525 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Nelumbo nucifera] |
| 3 |
Hb_001691_050 |
0.2069699737 |
- |
- |
- |
| 4 |
Hb_040506_010 |
0.2339506983 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000028_630 |
0.2405541489 |
- |
- |
PIN-like protein, partial [Selaginella apoda] |
| 6 |
Hb_001903_080 |
0.2443713284 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 7 |
Hb_000733_040 |
0.2476648291 |
- |
- |
hypothetical protein POPTR_0010s20310g [Populus trichocarpa] |
| 8 |
Hb_001178_120 |
0.2492944979 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 9 |
Hb_000395_100 |
0.2542378045 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 10 |
Hb_129067_010 |
0.2609906901 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 11 |
Hb_158990_010 |
0.2623331166 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_032264_130 |
0.2647106315 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 13 |
Hb_001902_020 |
0.2660724005 |
- |
- |
- |
| 14 |
Hb_000605_010 |
0.277596888 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 15 |
Hb_073212_010 |
0.2783521746 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 16 |
Hb_000153_020 |
0.2831543432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_088542_010 |
0.2839192057 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 18 |
Hb_000186_100 |
0.284670514 |
- |
- |
PREDICTED: uncharacterized protein LOC105649284 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008707_030 |
0.2851382285 |
- |
- |
PREDICTED: uncharacterized protein LOC104447982 [Eucalyptus grandis] |
| 20 |
Hb_007634_020 |
0.2860109635 |
- |
- |
- |