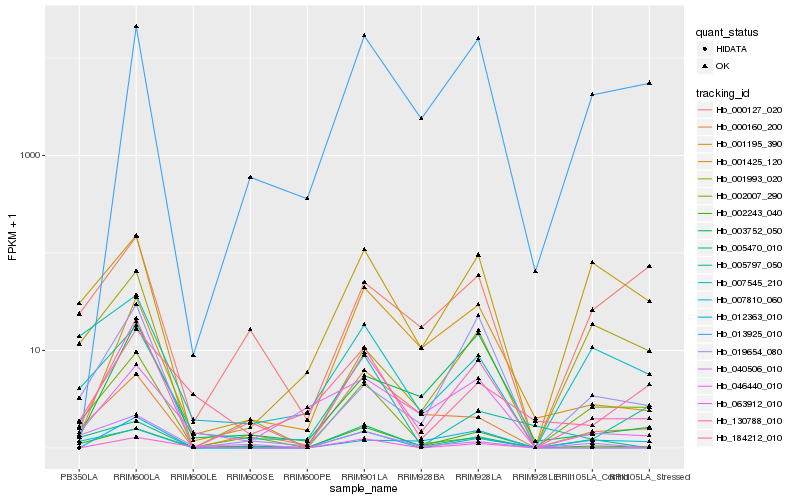
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130788_010 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 2 |
Hb_002243_040 |
0.2202770761 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 3 |
Hb_013925_010 |
0.2387423835 |
- |
- |
RecName: Full=Pro-hevein; AltName: Full=Major hevein; Contains: RecName: Full=Hevein; AltName: Allergen=Hev b 6; Contains: RecName: Full=Win-like protein; Flags: Precursor [Hevea brasiliensis] |
| 4 |
Hb_003752_050 |
0.2409603414 |
- |
- |
hypothetical protein JCGZ_15078 [Jatropha curcas] |
| 5 |
Hb_019654_080 |
0.2616555324 |
- |
- |
- |
| 6 |
Hb_005470_010 |
0.2714550915 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000160_200 |
0.274346082 |
- |
- |
PREDICTED: uncharacterized protein LOC105648226 [Jatropha curcas] |
| 8 |
Hb_001195_390 |
0.279414511 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |
| 9 |
Hb_040506_010 |
0.2928538825 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001425_120 |
0.2942732389 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 11 |
Hb_063912_010 |
0.2947079492 |
- |
- |
PREDICTED: elongation factor 1-alpha-like, partial [Cucumis sativus] |
| 12 |
Hb_005797_050 |
0.2948464684 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 13 |
Hb_001993_020 |
0.2966833647 |
- |
- |
hypothetical protein JCGZ_02641 [Jatropha curcas] |
| 14 |
Hb_046440_010 |
0.2967324552 |
- |
- |
hypothetical protein JCGZ_06190 [Jatropha curcas] |
| 15 |
Hb_012363_010 |
0.2990382003 |
- |
- |
- |
| 16 |
Hb_000127_020 |
0.3009106604 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_184212_010 |
0.303490922 |
- |
- |
PREDICTED: uncharacterized protein LOC105113658 [Populus euphratica] |
| 18 |
Hb_007810_060 |
0.3051692893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002007_290 |
0.3073528871 |
- |
- |
- |
| 20 |
Hb_007545_210 |
0.3079832724 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |