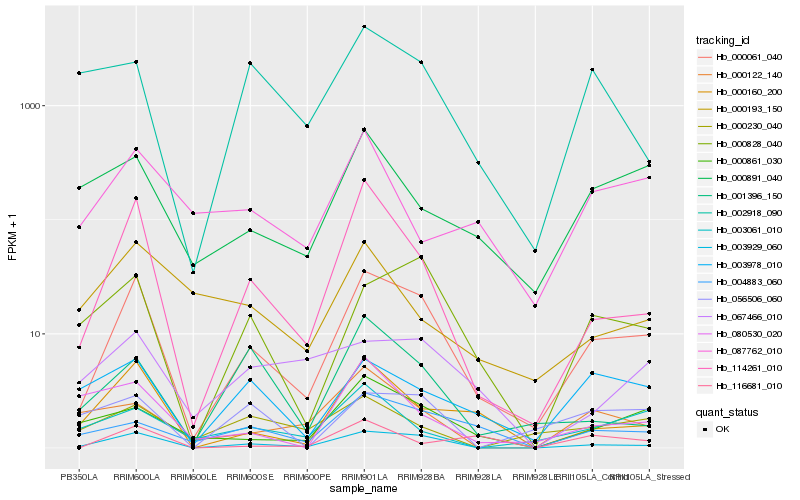
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_040 |
0.0 |
- |
- |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 2 |
Hb_003929_060 |
0.1409777156 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 3 |
Hb_114261_010 |
0.2041969631 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 4 |
Hb_000160_200 |
0.2397895826 |
- |
- |
PREDICTED: uncharacterized protein LOC105648226 [Jatropha curcas] |
| 5 |
Hb_000828_040 |
0.2400847243 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_000861_030 |
0.2539761908 |
- |
- |
- |
| 7 |
Hb_004883_060 |
0.2589324495 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003978_010 |
0.2666904265 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000891_040 |
0.2790140061 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 10 |
Hb_001396_150 |
0.2890058715 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 11 |
Hb_003061_010 |
0.2905810688 |
- |
- |
hypothetical protein SORBIDRAFT_01g039870 [Sorghum bicolor] |
| 12 |
Hb_056506_060 |
0.2936325273 |
- |
- |
hypothetical protein B456_007G294000 [Gossypium raimondii] |
| 13 |
Hb_002918_090 |
0.2954911327 |
- |
- |
PREDICTED: auxin-repressed 12.5 kDa protein-like [Nelumbo nucifera] |
| 14 |
Hb_087762_010 |
0.2959514379 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 15 |
Hb_000122_140 |
0.2963514254 |
- |
- |
- |
| 16 |
Hb_067466_010 |
0.2973034777 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 17 |
Hb_000230_040 |
0.2977896378 |
- |
- |
PREDICTED: protein MON2 homolog isoform X2 [Vitis vinifera] |
| 18 |
Hb_000193_150 |
0.3067081486 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 19 |
Hb_080530_020 |
0.3073895369 |
- |
- |
- |
| 20 |
Hb_116681_010 |
0.3083727038 |
- |
- |
hypothetical protein PRUPE_ppa025968mg, partial [Prunus persica] |