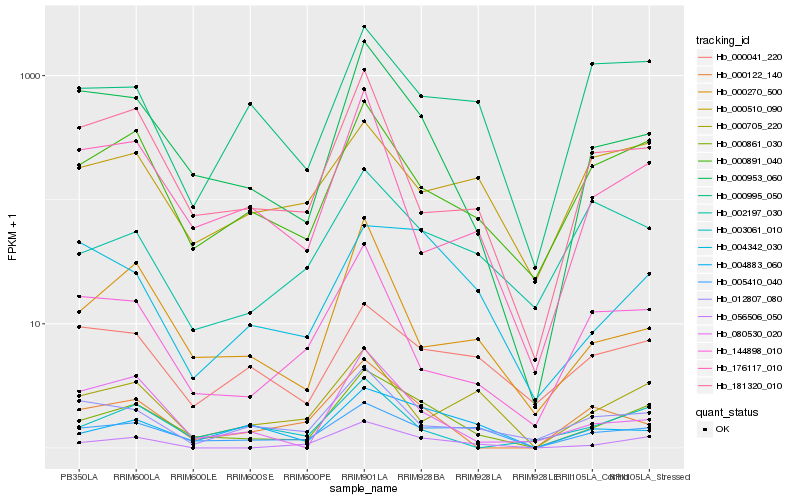
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000861_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000953_060 |
0.1670965138 |
- |
- |
PREDICTED: putative phytosulfokines 6 [Vitis vinifera] |
| 3 |
Hb_005410_040 |
0.1739867596 |
- |
- |
PREDICTED: probable ADP,ATP carrier protein At5g56450 [Jatropha curcas] |
| 4 |
Hb_000891_040 |
0.1772865987 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 5 |
Hb_056506_050 |
0.1779722121 |
- |
- |
hypothetical protein B456_007G294000 [Gossypium raimondii] |
| 6 |
Hb_004883_060 |
0.2060391856 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004342_030 |
0.2223309335 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 8 |
Hb_181320_010 |
0.2258323982 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 9 |
Hb_000122_140 |
0.2268170711 |
- |
- |
- |
| 10 |
Hb_000995_050 |
0.230285172 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 11 |
Hb_000270_500 |
0.2314546536 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 12 |
Hb_000705_220 |
0.2321878658 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_012807_080 |
0.2326013797 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 14 |
Hb_000041_220 |
0.2333366791 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 15 |
Hb_176117_010 |
0.2358441344 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 16 |
Hb_003061_010 |
0.2362614453 |
- |
- |
hypothetical protein SORBIDRAFT_01g039870 [Sorghum bicolor] |
| 17 |
Hb_002197_030 |
0.239020406 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_080530_020 |
0.2403562114 |
- |
- |
- |
| 19 |
Hb_144898_010 |
0.2405581379 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 20 |
Hb_000510_090 |
0.2442262825 |
- |
- |
40S ribosomal protein S15 [Morus notabilis] |