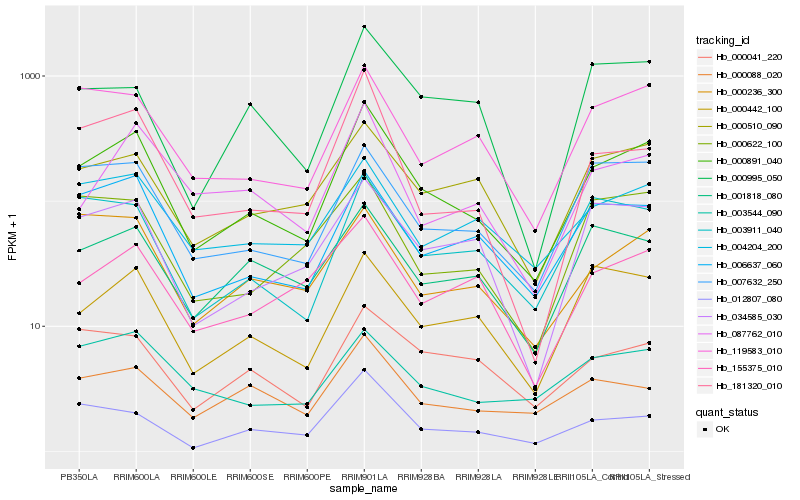
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000891_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 2 |
Hb_006637_060 |
0.1339093342 |
- |
- |
PREDICTED: uncharacterized protein LOC105649300 [Jatropha curcas] |
| 3 |
Hb_007632_250 |
0.1348910796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_119583_010 |
0.1361212965 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 5 |
Hb_000510_090 |
0.1370561032 |
- |
- |
40S ribosomal protein S15 [Morus notabilis] |
| 6 |
Hb_000442_100 |
0.1393831147 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 7 |
Hb_181320_010 |
0.1410156499 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 8 |
Hb_034585_030 |
0.1420700811 |
transcription factor |
TF Family: C2C2-Dof |
dof zinc finger protein DOF3.4 [Jatropha curcas] |
| 9 |
Hb_003911_040 |
0.1466432091 |
- |
- |
PREDICTED: putative H/ACA ribonucleoprotein complex subunit 1-like protein 1 [Jatropha curcas] |
| 10 |
Hb_012807_080 |
0.1467664996 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 11 |
Hb_004204_200 |
0.1483125762 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 12 |
Hb_087762_010 |
0.1499727714 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 13 |
Hb_001818_080 |
0.1512514276 |
- |
- |
PREDICTED: F-box protein At3g58530 isoform X1 [Jatropha curcas] |
| 14 |
Hb_155375_010 |
0.1516892718 |
- |
- |
hypothetical protein JCGZ_23388 [Jatropha curcas] |
| 15 |
Hb_000041_220 |
0.1548686718 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 16 |
Hb_003544_090 |
0.1568784154 |
- |
- |
PREDICTED: protein HIRA isoform X2 [Nelumbo nucifera] |
| 17 |
Hb_000236_300 |
0.1571025961 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Jatropha curcas] |
| 18 |
Hb_000088_020 |
0.1575619436 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 19 |
Hb_000995_050 |
0.157569299 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 20 |
Hb_000622_100 |
0.1584712307 |
- |
- |
PREDICTED: pre-rRNA-processing protein RIX1-like [Jatropha curcas] |