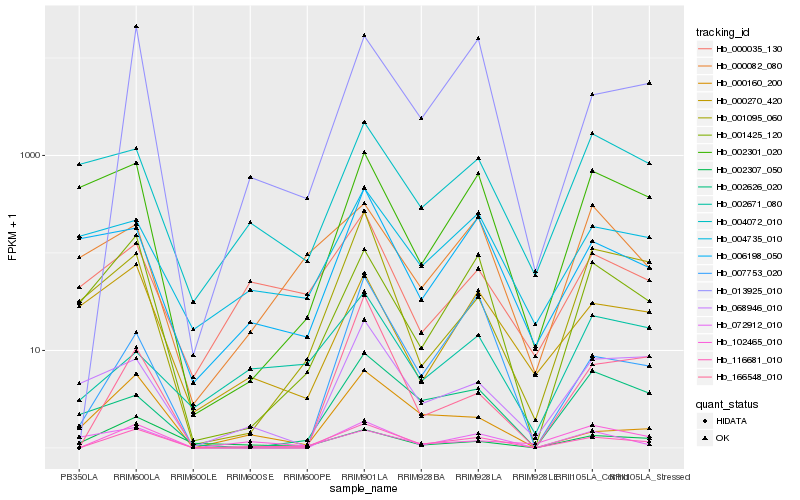
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116681_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa025968mg, partial [Prunus persica] |
| 2 |
Hb_013925_010 |
0.1802468091 |
- |
- |
RecName: Full=Pro-hevein; AltName: Full=Major hevein; Contains: RecName: Full=Hevein; AltName: Allergen=Hev b 6; Contains: RecName: Full=Win-like protein; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_001425_120 |
0.2375813981 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 4 |
Hb_000160_200 |
0.2504339236 |
- |
- |
PREDICTED: uncharacterized protein LOC105648226 [Jatropha curcas] |
| 5 |
Hb_000035_130 |
0.2587615207 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 6 |
Hb_102465_010 |
0.2609958922 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 7 |
Hb_001095_060 |
0.2649334815 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 8 |
Hb_166548_010 |
0.2658147606 |
- |
- |
hypothetical protein JCGZ_09681 [Jatropha curcas] |
| 9 |
Hb_068946_010 |
0.2693164453 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 10 |
Hb_002626_020 |
0.2708289399 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 11 |
Hb_004735_010 |
0.2747014891 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 12 |
Hb_007753_020 |
0.2796384287 |
- |
- |
- |
| 13 |
Hb_006198_050 |
0.2807547773 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000270_420 |
0.281363913 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 15 |
Hb_002301_020 |
0.2814937447 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 16 |
Hb_004072_010 |
0.2815582297 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_072912_010 |
0.2817076422 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 18 |
Hb_002307_050 |
0.2824473206 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000082_080 |
0.2838348016 |
- |
- |
putative polyol transporter protein 1 [Hevea brasiliensis] |
| 20 |
Hb_002671_080 |
0.2840268456 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10-like isoform X1 [Jatropha curcas] |