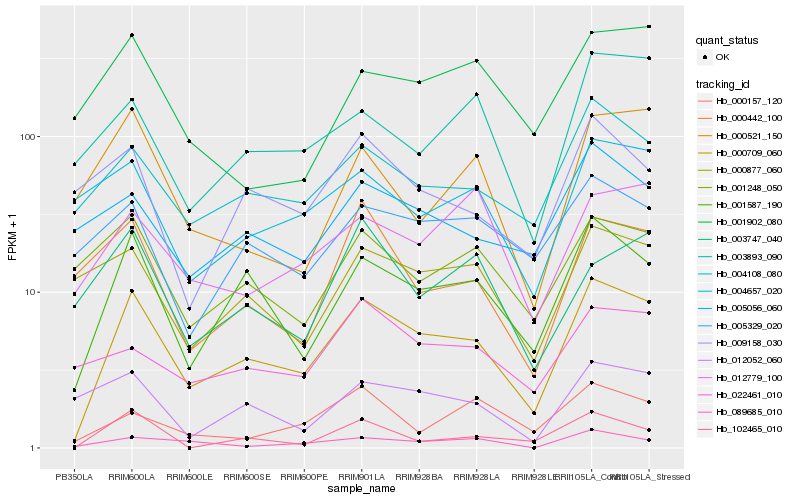
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000709_060 |
0.0 |
- |
- |
PREDICTED: abscisic-aldehyde oxidase-like isoform X5 [Populus euphratica] |
| 2 |
Hb_001587_190 |
0.1250816126 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 3 |
Hb_004108_080 |
0.1756899874 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase [Vitis vinifera] |
| 4 |
Hb_003893_090 |
0.1987073622 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 5 |
Hb_102465_010 |
0.2002307986 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 6 |
Hb_089685_010 |
0.2015945186 |
- |
- |
- |
| 7 |
Hb_000442_100 |
0.2027317168 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 8 |
Hb_005056_060 |
0.2034285639 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 9 |
Hb_001248_050 |
0.2041435171 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 10 |
Hb_000877_060 |
0.2052453516 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 11 |
Hb_004657_020 |
0.2054068278 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 12 |
Hb_005329_020 |
0.2064422951 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 13 |
Hb_022461_010 |
0.2069021908 |
- |
- |
- |
| 14 |
Hb_012779_100 |
0.2069942808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012052_060 |
0.2073262814 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 16 |
Hb_009158_030 |
0.2084865174 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_000521_150 |
0.2091235137 |
- |
- |
PREDICTED: uncharacterized protein LOC105650332 [Jatropha curcas] |
| 18 |
Hb_000157_120 |
0.2099974836 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_003747_040 |
0.2122727611 |
- |
- |
- |
| 20 |
Hb_001902_080 |
0.213160102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |