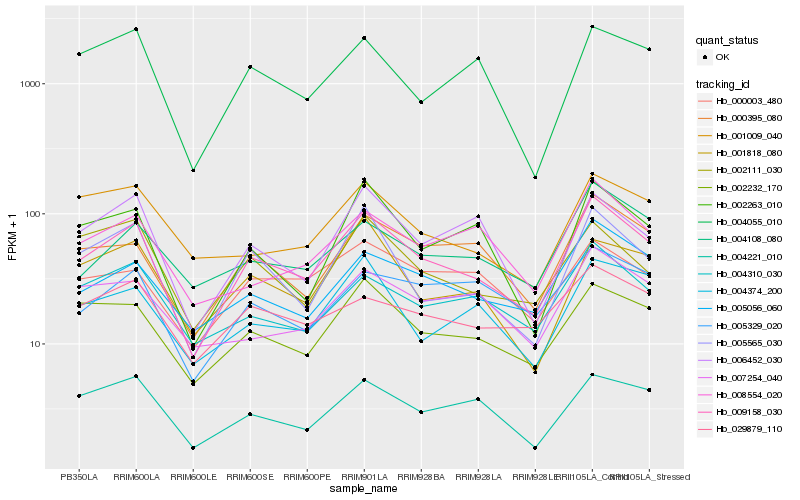
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009158_030 |
0.0 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 2 |
Hb_005056_060 |
0.104338319 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 3 |
Hb_002232_170 |
0.106743725 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 4 |
Hb_000395_080 |
0.106964012 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005565_030 |
0.1087786405 |
- |
- |
PREDICTED: uncharacterized protein LOC105633107 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004221_010 |
0.118039907 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 7 |
Hb_004310_030 |
0.1204317742 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 8 |
Hb_004108_080 |
0.1216014242 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase [Vitis vinifera] |
| 9 |
Hb_002263_010 |
0.1229867049 |
- |
- |
hypothetical protein CICLE_v10012259mg [Citrus clementina] |
| 10 |
Hb_004374_200 |
0.1238991812 |
- |
- |
putative GMP synthase [Oxybasis rubra] |
| 11 |
Hb_001818_080 |
0.125063047 |
- |
- |
PREDICTED: F-box protein At3g58530 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008554_020 |
0.1275128508 |
- |
- |
- |
| 13 |
Hb_006452_030 |
0.1284107965 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002111_030 |
0.1289314485 |
- |
- |
PREDICTED: NEDD8 ultimate buster 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_029879_110 |
0.1291809899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000003_480 |
0.1299847237 |
- |
- |
PREDICTED: nuclear pore complex protein NUP35 [Jatropha curcas] |
| 17 |
Hb_005329_020 |
0.1302831586 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 18 |
Hb_001009_040 |
0.1306024729 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Jatropha curcas] |
| 19 |
Hb_007254_040 |
0.1320208306 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004055_010 |
0.1326678809 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |