| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
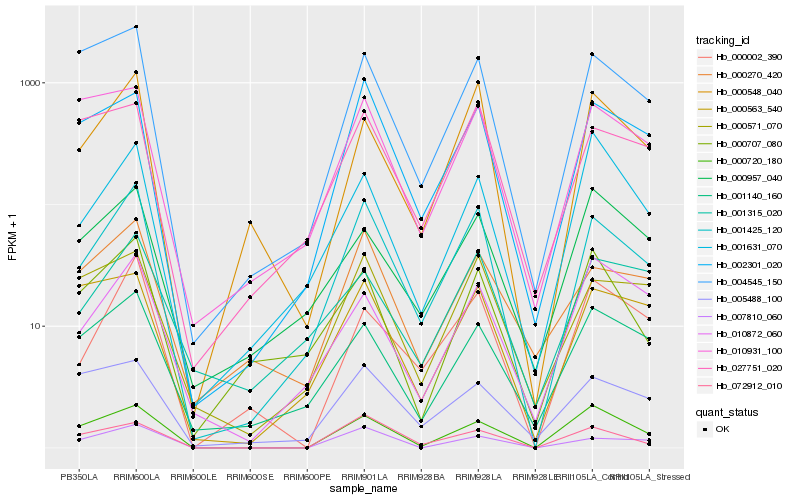
Hb_001425_120 |
0.0 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 2 |
Hb_002301_020 |
0.1393943369 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 3 |
Hb_000548_040 |
0.1482081302 |
rubber biosynthesis |
Gene Name: CPT1 |
cis-prenyltransferase1 [Hevea brasiliensis] |
| 4 |
Hb_004545_150 |
0.1628420915 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 5 |
Hb_000270_420 |
0.163540389 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 6 |
Hb_007810_060 |
0.1638407086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000571_070 |
0.1643077282 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 8 |
Hb_000720_180 |
0.1650351319 |
- |
- |
- |
| 9 |
Hb_001140_160 |
0.166679696 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 10 |
Hb_000002_390 |
0.1678892138 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_072912_010 |
0.1682576394 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 12 |
Hb_027751_020 |
0.1715122731 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_010931_100 |
0.1781701595 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 14 |
Hb_010872_060 |
0.1786240667 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 15 |
Hb_000707_080 |
0.1796182956 |
- |
- |
hypothetical protein AALP_AA4G156500, partial [Arabis alpina] |
| 16 |
Hb_000563_540 |
0.1799803294 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 17 |
Hb_001315_020 |
0.1813393266 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_001631_070 |
0.1820987789 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 19 |
Hb_005488_100 |
0.183243168 |
- |
- |
hypothetical protein JCGZ_00295 [Jatropha curcas] |
| 20 |
Hb_000957_040 |
0.1850480213 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |