| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
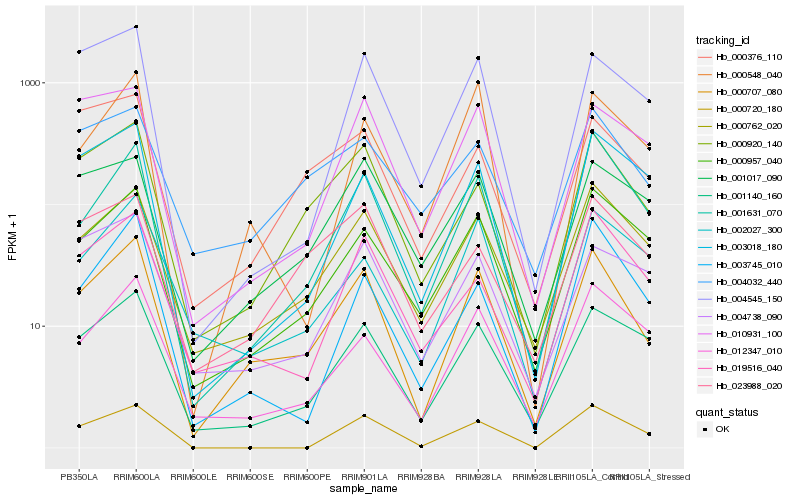
Hb_000707_080 |
0.0 |
- |
- |
hypothetical protein AALP_AA4G156500, partial [Arabis alpina] |
| 2 |
Hb_000762_020 |
0.1305087687 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 3 |
Hb_000920_140 |
0.1396915917 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_001140_160 |
0.1436707677 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_001631_070 |
0.1471071851 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000957_040 |
0.148170357 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 7 |
Hb_012347_010 |
0.1589955456 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 8 |
Hb_002027_300 |
0.1638851413 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 9 |
Hb_000548_040 |
0.165254336 |
rubber biosynthesis |
Gene Name: CPT1 |
cis-prenyltransferase1 [Hevea brasiliensis] |
| 10 |
Hb_019516_040 |
0.1652931013 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 11 |
Hb_001017_090 |
0.1681276622 |
- |
- |
- |
| 12 |
Hb_004032_440 |
0.1691233968 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 13 |
Hb_003018_180 |
0.1714626901 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 14 |
Hb_004545_150 |
0.1735122072 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 15 |
Hb_010931_100 |
0.17380457 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 16 |
Hb_004738_090 |
0.1745671067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023988_020 |
0.1751642719 |
- |
- |
PREDICTED: transmembrane protein 19 [Prunus mume] |
| 18 |
Hb_000376_110 |
0.1752733408 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Brachypodium distachyon] |
| 19 |
Hb_000720_180 |
0.178226647 |
- |
- |
- |
| 20 |
Hb_003745_010 |
0.1784565155 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |