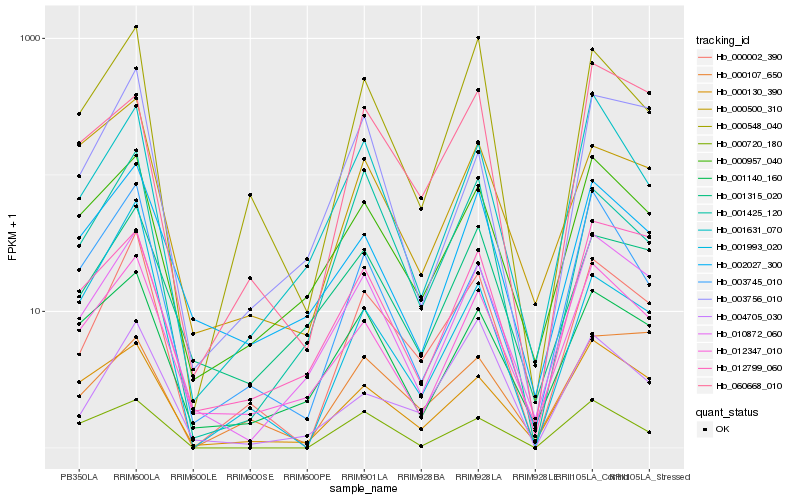
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_390 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000548_040 |
0.1271831231 |
rubber biosynthesis |
Gene Name: CPT1 |
cis-prenyltransferase1 [Hevea brasiliensis] |
| 3 |
Hb_010872_060 |
0.1667351295 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 4 |
Hb_001425_120 |
0.1678892138 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 5 |
Hb_012347_010 |
0.1724926051 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 6 |
Hb_000957_040 |
0.1769476655 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 7 |
Hb_001631_070 |
0.1780028248 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 8 |
Hb_000130_390 |
0.1792120299 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 9 |
Hb_003745_010 |
0.1793036186 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 10 |
Hb_004705_030 |
0.183622677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003756_010 |
0.1874956811 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 12 |
Hb_002027_300 |
0.1885407657 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 13 |
Hb_001993_020 |
0.1887608902 |
- |
- |
hypothetical protein JCGZ_02641 [Jatropha curcas] |
| 14 |
Hb_001315_020 |
0.1912198534 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_001140_160 |
0.1920275419 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 16 |
Hb_000720_180 |
0.1935790744 |
- |
- |
- |
| 17 |
Hb_000107_650 |
0.1988944842 |
- |
- |
PREDICTED: cyclic phosphodiesterase-like [Jatropha curcas] |
| 18 |
Hb_060668_010 |
0.1998428706 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 19 |
Hb_012799_060 |
0.2034565039 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 20 |
Hb_000500_310 |
0.2054612807 |
- |
- |
- |