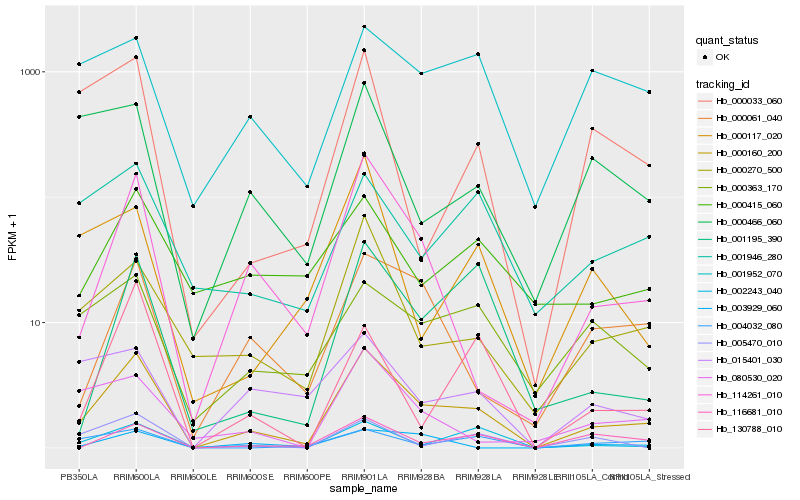
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648226 [Jatropha curcas] |
| 2 |
Hb_114261_010 |
0.2133273238 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 3 |
Hb_000270_500 |
0.2234886347 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 4 |
Hb_002243_040 |
0.236345134 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 5 |
Hb_000061_040 |
0.2397895826 |
- |
- |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 6 |
Hb_116681_010 |
0.2504339236 |
- |
- |
hypothetical protein PRUPE_ppa025968mg, partial [Prunus persica] |
| 7 |
Hb_004032_080 |
0.2538401194 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL11 [Jatropha curcas] |
| 8 |
Hb_001195_390 |
0.25617444 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |
| 9 |
Hb_080530_020 |
0.2617323337 |
- |
- |
- |
| 10 |
Hb_000033_060 |
0.2621645091 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 11 |
Hb_000363_170 |
0.2652319477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000466_060 |
0.2656545848 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 13 |
Hb_001946_280 |
0.2712506392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001952_070 |
0.2714989106 |
- |
- |
40S ribosomal protein S6B [Hevea brasiliensis] |
| 15 |
Hb_005470_010 |
0.2715862184 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_015401_030 |
0.2719706874 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_000415_060 |
0.2728432722 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 18 |
Hb_130788_010 |
0.274346082 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 19 |
Hb_003929_060 |
0.2754220819 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 20 |
Hb_000117_020 |
0.2772932386 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |