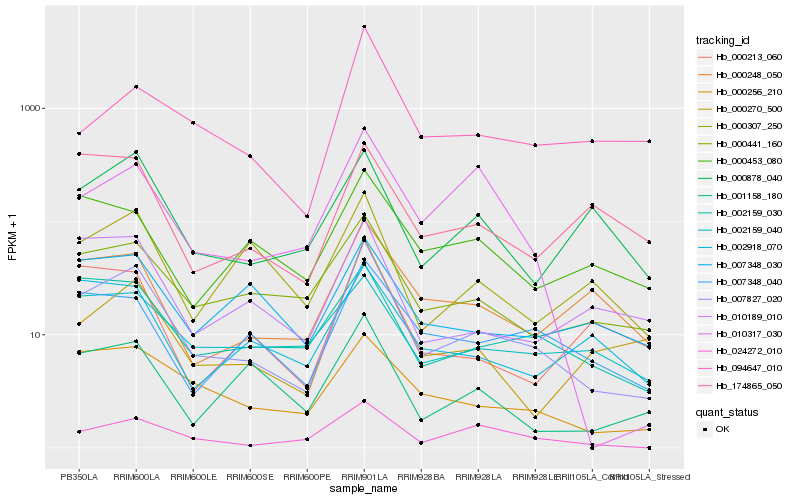
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007827_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000256_210 |
0.1722100103 |
- |
- |
PREDICTED: 40S ribosomal protein S11-like [Phoenix dactylifera] |
| 3 |
Hb_002159_030 |
0.1742616401 |
- |
- |
AP-1 complex subunit gamma-2, putative [Ricinus communis] |
| 4 |
Hb_000441_160 |
0.1765415032 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 5 |
Hb_024272_010 |
0.1826098384 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_007348_040 |
0.1855194494 |
transcription factor |
TF Family: PHD |
hypothetical protein RCOM_1314010 [Ricinus communis] |
| 7 |
Hb_000270_500 |
0.1893990251 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 8 |
Hb_010189_010 |
0.1894586101 |
- |
- |
- |
| 9 |
Hb_000248_050 |
0.1928883607 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001158_180 |
0.1996459533 |
- |
- |
- |
| 11 |
Hb_000307_250 |
0.2032985563 |
- |
- |
PREDICTED: probable nucleoredoxin 2 [Jatropha curcas] |
| 12 |
Hb_000878_040 |
0.2034137923 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002159_040 |
0.2060727747 |
- |
- |
hypothetical protein PRUPE_ppa001563mg [Prunus persica] |
| 14 |
Hb_002918_070 |
0.2079735948 |
- |
- |
hypothetical protein POPTR_0011s05530g [Populus trichocarpa] |
| 15 |
Hb_094647_010 |
0.2085527926 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 16 |
Hb_007348_030 |
0.2097102097 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000213_060 |
0.210100649 |
- |
- |
PREDICTED: RRP12-like protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_010317_030 |
0.2107089748 |
- |
- |
unknown [Picea sitchensis] |
| 19 |
Hb_000453_080 |
0.2140188698 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_174865_050 |
0.2155256734 |
- |
- |
PREDICTED: neuroguidin [Jatropha curcas] |