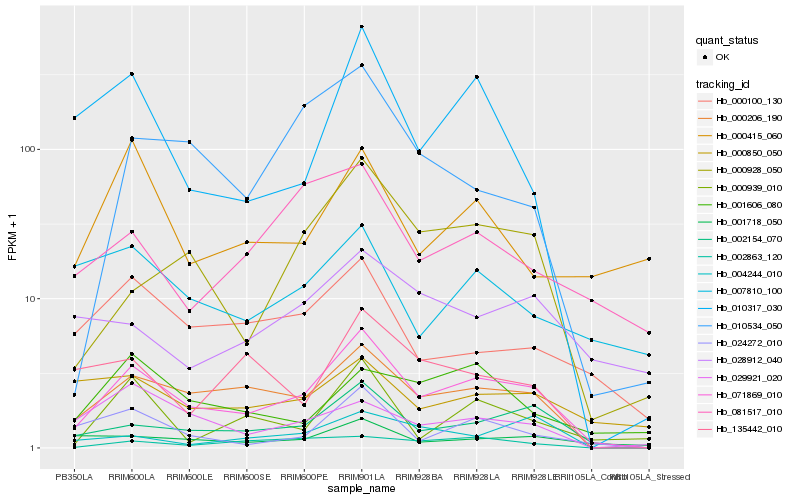
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071869_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000206_190 |
0.1444974011 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 3 |
Hb_002154_070 |
0.1590418177 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 4 |
Hb_000928_050 |
0.2084050984 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 5 |
Hb_024272_010 |
0.2136639634 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_010317_030 |
0.2272651356 |
- |
- |
unknown [Picea sitchensis] |
| 7 |
Hb_000100_130 |
0.2286864827 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 8 |
Hb_002863_120 |
0.229979877 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 9 |
Hb_001718_050 |
0.2310234081 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Brassica rapa] |
| 10 |
Hb_010534_050 |
0.237859123 |
- |
- |
PREDICTED: 40S ribosomal protein S7 [Jatropha curcas] |
| 11 |
Hb_135442_010 |
0.241589662 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 12 |
Hb_000939_010 |
0.2425854426 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 13 |
Hb_081517_010 |
0.2450593126 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004244_010 |
0.2455864157 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 15 |
Hb_029921_020 |
0.2460211888 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001606_080 |
0.2498393031 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000850_050 |
0.253879442 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 18 |
Hb_007810_100 |
0.2554522665 |
- |
- |
Synaptic glycoprotein SC2, putative [Ricinus communis] |
| 19 |
Hb_028912_040 |
0.2567512815 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 20 |
Hb_000415_060 |
0.2572515028 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |