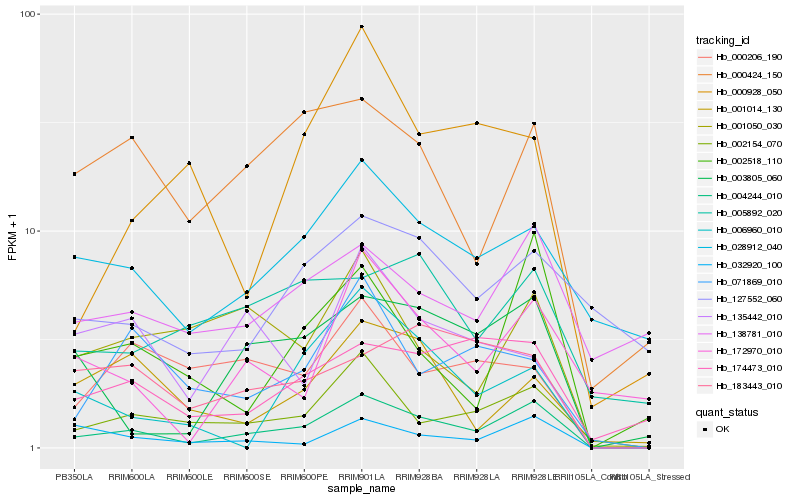
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004244_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 2 |
Hb_002154_070 |
0.1894752081 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 3 |
Hb_003805_060 |
0.2181099377 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_002518_110 |
0.2240637576 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase [Manihot esculenta] |
| 5 |
Hb_000424_150 |
0.2244073889 |
- |
- |
PREDICTED: copper transporter 5.1-like [Jatropha curcas] |
| 6 |
Hb_028912_040 |
0.2273110767 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 7 |
Hb_032920_100 |
0.2275311337 |
- |
- |
hypothetical protein POPTR_0002s00940g, partial [Populus trichocarpa] |
| 8 |
Hb_001050_030 |
0.2313856097 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 9 |
Hb_174473_010 |
0.2317978342 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 10 |
Hb_071869_010 |
0.2455864157 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 11 |
Hb_172970_010 |
0.2462082254 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 12 |
Hb_183443_010 |
0.2465859957 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 13 |
Hb_000206_190 |
0.2500872383 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 14 |
Hb_005892_020 |
0.2526660579 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 15 |
Hb_138781_010 |
0.2526953578 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 16 |
Hb_006960_010 |
0.2541067298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001014_130 |
0.2541775423 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 18 |
Hb_135442_010 |
0.2605116591 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 19 |
Hb_000928_050 |
0.2625640331 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 20 |
Hb_127552_060 |
0.2641199793 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
protein binding protein, putative [Ricinus communis] |