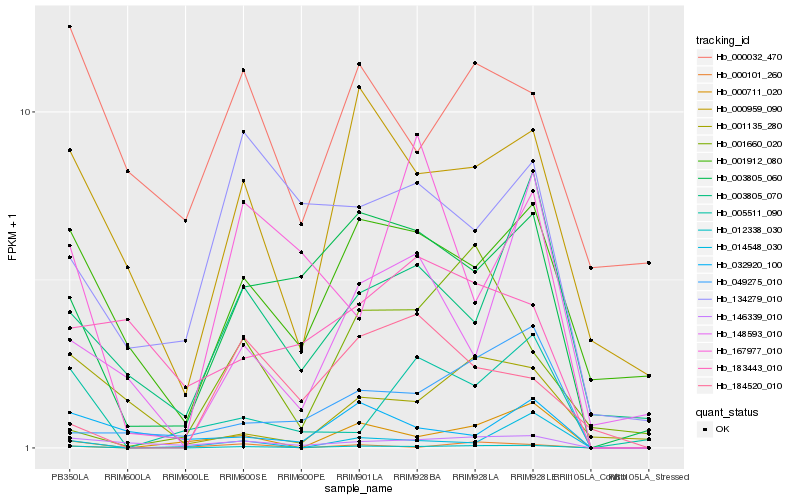
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012338_030 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_003805_060 |
0.2102925348 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_146339_010 |
0.2527023883 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 4 |
Hb_000959_090 |
0.2546451593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001912_080 |
0.2550772252 |
- |
- |
vacuolar protein sorting-associated protein, putative [Ricinus communis] |
| 6 |
Hb_003805_070 |
0.2787594517 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 7 |
Hb_000711_020 |
0.2887206188 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 8 |
Hb_049275_010 |
0.2896603371 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 9 |
Hb_000101_260 |
0.2906372483 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_005511_090 |
0.2917376787 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |
| 11 |
Hb_134279_010 |
0.30185847 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 12 |
Hb_167977_010 |
0.3033795976 |
- |
- |
hypothetical protein POPTR_0185s002102g, partial [Populus trichocarpa] |
| 13 |
Hb_184520_010 |
0.3111384406 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_000032_470 |
0.3170107077 |
- |
- |
PREDICTED: uncharacterized protein LOC103415826 [Malus domestica] |
| 15 |
Hb_014548_030 |
0.3202853435 |
- |
- |
PREDICTED: uncharacterized protein LOC104897580 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_148593_010 |
0.3211968267 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 17 |
Hb_032920_100 |
0.3238256293 |
- |
- |
hypothetical protein POPTR_0002s00940g, partial [Populus trichocarpa] |
| 18 |
Hb_001660_020 |
0.3279630813 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 19 |
Hb_183443_010 |
0.3283759402 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 20 |
Hb_001135_280 |
0.328918064 |
- |
- |
- |