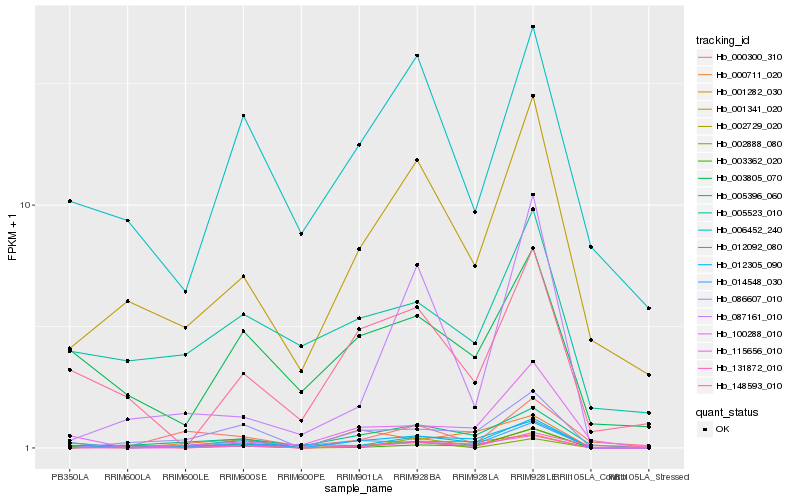
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012305_090 |
0.0 |
- |
- |
hypothetical protein VITISV_018166 [Vitis vinifera] |
| 2 |
Hb_086607_010 |
0.1702475328 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_005396_060 |
0.1715589649 |
- |
- |
- |
| 4 |
Hb_014548_030 |
0.1753936708 |
- |
- |
PREDICTED: uncharacterized protein LOC104897580 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001341_020 |
0.2130264863 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 6 |
Hb_000711_020 |
0.2163490904 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 7 |
Hb_115656_010 |
0.2473151866 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 8 |
Hb_148593_010 |
0.2498590437 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 9 |
Hb_003805_070 |
0.2505689211 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 10 |
Hb_100288_010 |
0.2507457131 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_002888_080 |
0.2512809862 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 12 |
Hb_002729_020 |
0.2583299724 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 13 |
Hb_003362_020 |
0.263707386 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 14 |
Hb_006452_240 |
0.2680980709 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 15 |
Hb_000300_310 |
0.2729307332 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_012092_080 |
0.2739001799 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 17 |
Hb_131872_010 |
0.2741751569 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_087161_010 |
0.2745686903 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_001282_030 |
0.2746106339 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 20 |
Hb_005523_010 |
0.2748714902 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |