| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
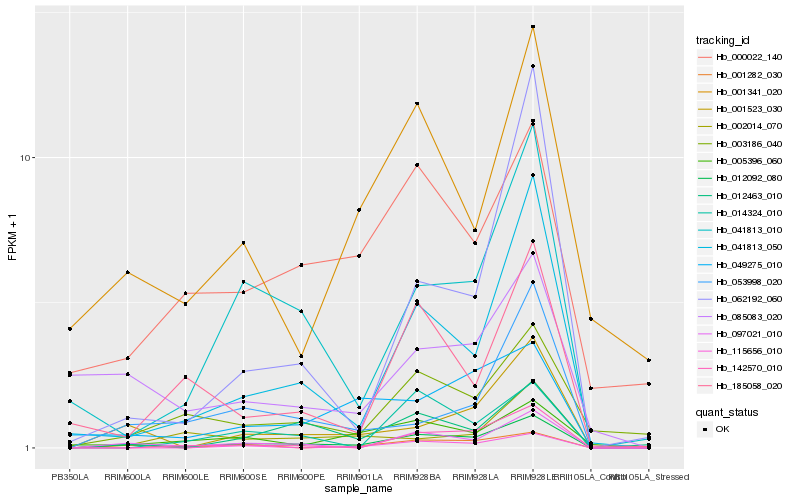
Hb_012092_080 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 2 |
Hb_097021_010 |
0.1568409363 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 3 |
Hb_041813_050 |
0.1806990151 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 4 |
Hb_001523_030 |
0.1837705945 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_012463_010 |
0.2047749923 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_001282_030 |
0.2070917525 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 7 |
Hb_041813_010 |
0.2141299339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_053998_020 |
0.2211702999 |
- |
- |
- |
| 9 |
Hb_142570_010 |
0.2280302718 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_049275_010 |
0.231695367 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 11 |
Hb_003186_040 |
0.2332521021 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 12 |
Hb_062192_060 |
0.2388410973 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 13 |
Hb_005396_060 |
0.2389363146 |
- |
- |
- |
| 14 |
Hb_115656_010 |
0.2392518368 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 15 |
Hb_185058_020 |
0.2459008763 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 16 |
Hb_001341_020 |
0.246026257 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 17 |
Hb_014324_010 |
0.2501605492 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 18 |
Hb_000022_140 |
0.2539531204 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 19 |
Hb_085083_020 |
0.2548028764 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 20 |
Hb_002014_070 |
0.2558206573 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |