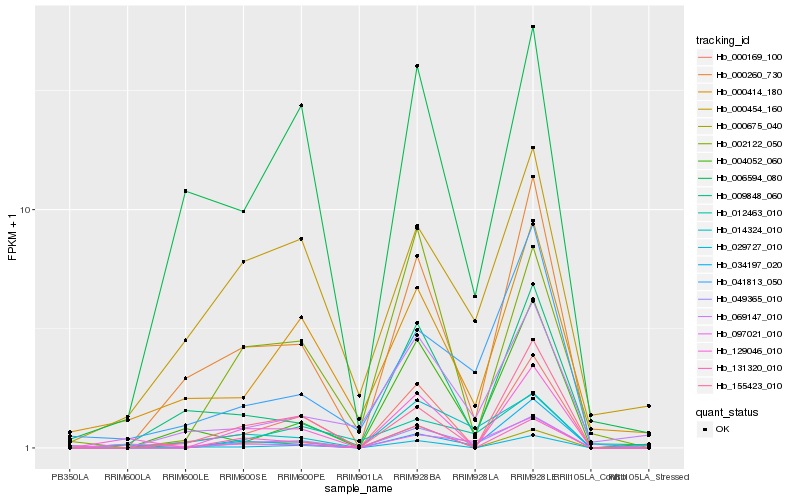
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049365_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 2 |
Hb_014324_010 |
0.2131958803 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 3 |
Hb_012463_010 |
0.2148016988 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_069147_010 |
0.2151650369 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 5 |
Hb_000260_730 |
0.2208160896 |
- |
- |
- |
| 6 |
Hb_131320_010 |
0.2256669617 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 7 |
Hb_034197_020 |
0.2312984004 |
- |
- |
hypothetical protein VITISV_041099 [Vitis vinifera] |
| 8 |
Hb_000169_100 |
0.234887021 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_004052_060 |
0.2394129195 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000454_160 |
0.2461753966 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000675_040 |
0.2464050083 |
- |
- |
hypothetical protein OsI_02937 [Oryza sativa Indica Group] |
| 12 |
Hb_129046_010 |
0.2473638455 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 13 |
Hb_002122_050 |
0.2484487486 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_009848_060 |
0.2497500603 |
- |
- |
unknown [Glycine max] |
| 15 |
Hb_029727_010 |
0.2513364126 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_000414_180 |
0.2549051198 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 17 |
Hb_041813_050 |
0.2583858364 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 18 |
Hb_155423_010 |
0.2606163187 |
- |
- |
- |
| 19 |
Hb_006594_080 |
0.2636490024 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_097021_010 |
0.2640330866 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |