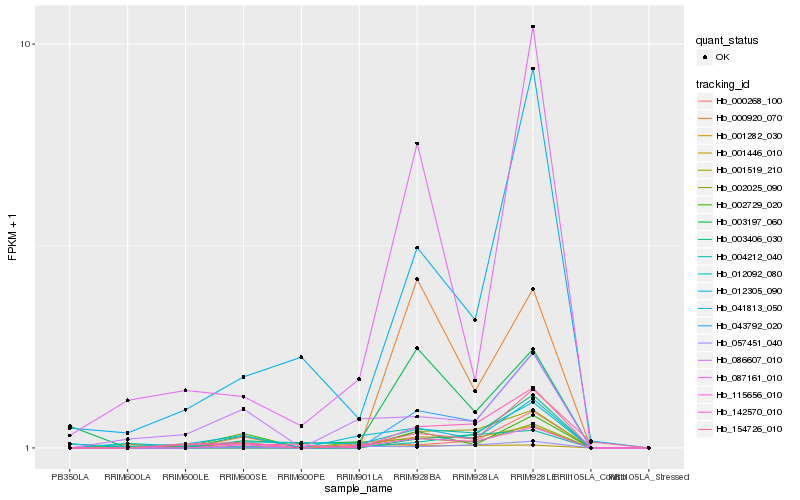
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115656_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 2 |
Hb_057451_040 |
0.1962684098 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001282_030 |
0.2013007224 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 4 |
Hb_004212_040 |
0.2117310847 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_002025_090 |
0.2325788888 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_012092_080 |
0.2392518368 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 7 |
Hb_142570_010 |
0.2401161602 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_012305_090 |
0.2473151866 |
- |
- |
hypothetical protein VITISV_018166 [Vitis vinifera] |
| 9 |
Hb_000268_100 |
0.2504863177 |
- |
- |
- |
| 10 |
Hb_043792_020 |
0.2547767705 |
- |
- |
Retrotransposon gag protein [Medicago truncatula] |
| 11 |
Hb_001519_210 |
0.2584770447 |
- |
- |
hypothetical protein B456_003G032500 [Gossypium raimondii] |
| 12 |
Hb_041813_050 |
0.262019362 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 13 |
Hb_000920_070 |
0.2650871618 |
- |
- |
- |
| 14 |
Hb_154726_010 |
0.2665951773 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 15 |
Hb_003406_030 |
0.2682746554 |
- |
- |
PREDICTED: uncharacterized protein LOC104774100 [Camelina sativa] |
| 16 |
Hb_002729_020 |
0.272567971 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 17 |
Hb_086607_010 |
0.2788587969 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_001446_010 |
0.2796286111 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_087161_010 |
0.2798554925 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_003197_060 |
0.2814876951 |
- |
- |
hypothetical protein POPTR_0015s11900g [Populus trichocarpa] |