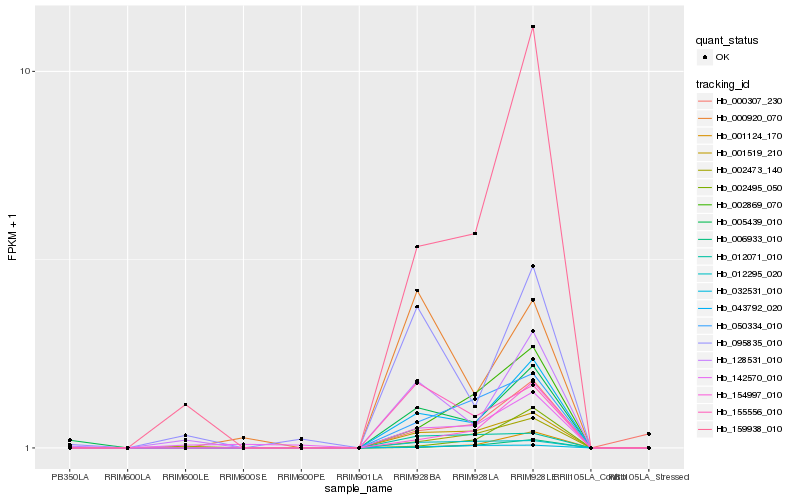
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043792_020 |
0.0 |
- |
- |
Retrotransposon gag protein [Medicago truncatula] |
| 2 |
Hb_001519_210 |
0.1109048889 |
- |
- |
hypothetical protein B456_003G032500 [Gossypium raimondii] |
| 3 |
Hb_159938_010 |
0.1220088083 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_005439_010 |
0.1526130296 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_154997_010 |
0.164126339 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 6 |
Hb_128531_010 |
0.1692407922 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 7 |
Hb_050334_010 |
0.1759202542 |
- |
- |
- |
| 8 |
Hb_002869_070 |
0.1853988481 |
- |
- |
- |
| 9 |
Hb_002473_140 |
0.1998596524 |
- |
- |
- |
| 10 |
Hb_142570_010 |
0.2132914722 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_095835_010 |
0.2153948628 |
- |
- |
hypothetical protein JCGZ_23410 [Jatropha curcas] |
| 12 |
Hb_155556_010 |
0.2171966035 |
- |
- |
putative methylenetetrahydrofolate reductase [Morus notabilis] |
| 13 |
Hb_006933_010 |
0.2225369691 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_012295_020 |
0.2407770647 |
- |
- |
PREDICTED: uncharacterized protein LOC104223093 [Nicotiana sylvestris] |
| 15 |
Hb_032531_010 |
0.243150302 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_012071_010 |
0.2434688556 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_000307_230 |
0.2470809244 |
- |
- |
- |
| 18 |
Hb_002495_050 |
0.2478710101 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 19 |
Hb_000920_070 |
0.2496014804 |
- |
- |
- |
| 20 |
Hb_001124_170 |
0.2500288686 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |